Abstract
Background
Histones chaperones have been found to play critical roles in tumor development and progression. However, the role of histone chaperone CHAF1A in gastric carcinogenesis and its underlying mechanisms remain elusive.
Methods
CHAF1A expression in gastric cancer (GC) was analyzed in GEO datasets and clinical specimens. CHAF1A knockdown and overexpression were used to explore its functions in gastric cancer cells. The regulation and potential molecular mechanism of CHAF1A expression in gastric cancer cells were studied by using cell and molecular biological methods.
Findings
CHAF1A was upregulated in GC tissues and its high expression predicted poor prognosis in GC patients. Overexpression of CHAF1A promoted gastric cancer cell proliferation both in vitro and in vivo, whereas CHAF1A suppression exhibited the opposite effects. Mechanistically, CHAF1A acted as a co-activator in the Wnt pathway. CHAF1A directly interacted with TCF4 to enhance the expression of c-MYC and CCND1 through binding to their promoter regions. In addition, the overexpression of CHAF1A was modulated by specificity protein 1 (Sp1) in GC. Sp1 transcriptionally enhanced the expression of CHAF1A in GC. Furthermore, CHAF1A expression induced by Helicobacter pylori was Sp1 dependent.
Interpretation
CHAF1A is a potential oncogene in GC, and may serve as a novel therapeutic target for GC treatment.
Keywords: Histone chaperone, CHAF1A, Gastric cancer, Carcinogenesis
Research in context.
Evidence before this study
Growing evidence has suggested that histone chaperones emerged as potential drivers in cancer initiation and progression. The histone chaperone CHAF1A has been found to play critical roles in the development and progression of solid tumors. However, the effects of CHAF1A on gastric carcinogenesis remain elusive.
Added value of this study
This study investigated the expression profile, biological function, downstream regulation, clinical impact of CHAF1A on gastric cancer (GC). We found that increased CHAF1A expression predicts poor clinical outcome in GC patients and promotes GC cell proliferation. CHAF1A directly interacts with TCF4 to enhance the expression of c-MYC and CCND1 in GC. In addition, our study first described that Helicobacter pylori can induce the expression of histone chaperone CHAF1A in specificity protein 1 (Sp1) dependent manner.
Implications of all the available evidence
These findings suggest that CHAF1A may serve as a potential target for the prevention and treatment of GC.
Alt-text: Unlabelled Box
1. Introduction
Gastric cancer (GC) is the third leading cause of cancer-related death worldwide and often results in a poor prognosis due to its late diagnosis [1]. However, despite significant advances in modern medicine over the past century, there has been little improvement in the treatment of GC. The majority of GC patients are diagnosed at an advanced stage, which leads to poor prognosis and 5-year overall survival rate [2,3]. Therefore, it is necessary to elucidate the molecular mechanisms of GC and explore the potential diagnostic, prognostic and therapeutic biomarkers for GC patients.
The complexity of carcinogenesis is not only caused by genetic alterations, but also involves epigenetic modifications. The major epigenetic features of cancer cells include DNA methylation, histone modifications and non-coding RNAs, which can alter the expression of cancer-related gene [4,5]. Recently, it has been reported that epigenetic alterations, such as promoter CpG methylation and histone modification enzymes, are involved in the development and progression of GC [[6], [7], [8]]. Growing evidence has suggested that histone variants and their chaperones emerged as potential drivers in cancer initiation and progression [9,10].
Chromatin assembly factor-1 (CAF-1) is a highly conserved histone chaperone heterotrimer, which consists of p48, p60 and p150 (CHAF1A) subunits. CAF-1 plays an essential role in diverse biological processes, such as DNA replication during the nucleosome formation and the chromatin restoration after DNA repair [[11], [12], [13], [14], [15]]. As a core component of CAF-1, CHAF1A epigenetically regulates gene expression by interacting with heterochromatin protein 1 (HP1) [16,17]. In addition, CHAF1A participates in a complex with methyl CpG DNA binding domain protein 1 (MDB1) and histone methyl transferase SETDB1 during initiation of a gene-silencing program by promoting H3K9 trimethylation, heterochromatin formation, and DNA methylation [18,19]. CHAF1A enhances Gfi1-mediated transcriptional repression and occupies Gfi1 target gene promoters in transfected cells [20]. Recently, CHAF1A has been associated with the development and progression of solid tumors, including breast cancer, prostate squamous cell carcinoma, hepatocellular carcinoma, glioma and neuroblastoma [[21], [22], [23], [24], [25], [26], [27]]. However, the role of CHAF1A in GC remains largely unknown. Therefore, in this study, we aimed to investigate the expression profile, biological function, downstream regulation, clinical impact of CHAF1A on GC.
The sustained and uncontrolled cellular growth is one of the hallmarks of cancer cells [28]. Several signaling pathways, such as Wnt pathway, can determine the growth of tumor cells [[29], [30], [31]]. Aberrant activation of Wnt pathway plays a central role in the oncogenic processes of GC [32,33]. However, it remains unclear whether CHAF1A contributes to the regulation of Wnt pathway. Here, we elucidate the molecular mechanisms linking CHAF1A and Wnt pathway.
2. Materials and methods
2.1. Cell culture
GC cell lines BGC-823, HGC-27, MGC-803 and SGC-7901 were cultured in RPMI-1640 (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco, Carlsbad, CA, USA). AGS cells were cultured in F12 (HyClone, USA) containing 10% FBS. BGC-823 cells with stably over-expression of CHAF1A were selected using 3 μg/mL puromycin (Gibco, Carlsbad, CA, USA). All cultures were maintained in a humidified 5% CO2 incubator at 37 °C.
2.2. siRNA and plasmids transfection
CHAF1A and Sp1 siRNAs (Sigma-Aldrich, USA) were transfected into GC cells by lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Flag-tagged CHAF1A (Genechem, Shanghai, China) and Myc-tagged TCF4 (Genechem, Shanghai, China) were transfected with Roche Transfection Reagent (Roche, Switzerland). Sequences for these siRNAs are listed in Table S1.
2.3. Human clinical specimens
Thirty six RNA samples of GC and adjacent non-tumor tissues were collected from Shandong Tumor Hospital, while 91 RNA samples of atrophic gastritis (AG) with H. pylori positive or negative were obtained from Jinan Central Hospital, Shandong, P. R. China. The diagnosis of H. pylori infection in AG patients was performed by 13C urea breath test. For protein samples, five pairs of GC and matched adjacent non-tumor gastric tissues were frozen and stored in liquid nitrogen until further analysis. In addition, FFPE samples of AG (n = 39), GC tissues (n = 35) and adjacent non-tumor tissues (n = 35) were collected immediately after endoscopic biopsy or surgery and then stored in formalin. Histological examination was used to confirm the diagnosis in all cases. There was no relation between RNA samples and FFPE samples. The study protocol was approved by Shandong University Research Ethics Committee.
2.4. H. pylori cultures
H. pylori strains 26695 and 11637 were grown in Brucella broth supplemented with 5% FBS under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) at 37 °C. The bacteria were harvested by centrifugation, and immediately transferred to cell cultures at a multiplicity of infection (MOI) of 100.
2.5. Colony formation and cell viability assay
Cells (500 cells/well) were seeded in 6-well plate and incubated for 8–14 days until colonies of cells appeared. The colonies were fixed with methanol and stained with Giemsa. Short-term cell proliferation was examined using cell counting kit-8 (CCK-8) assay (MedChemExpress, USA). In brief, cells were seeded at 1000 cells per well (100 μL) in a 96-well plate. Afterwards, 10 μL of CCK-8 solution was added into each well and incubated for 2 h to determine the cell viability. The optical density was measured using a microplate reader at 450 nm (reference wavelength 630 nm). All experiments were performed in triplicate.
2.6. Immunohistochemistry
FFPE tissue sections acquired from patients were subjected to deparaffinization and dehydration. After antigen retrieval, H2O2 treatment and non-specific antigens blocking, the slides were incubated with monoclonal rabbit anti-human CHAF1A (1:100, Abcam, ab126625) at 4 °C. After overnight incubation, the slides were incubated with secondary antibody, followed by colorimetric detection using DAB staining kit (Vector Laboratories, USA).
2.7. RNA extraction, RT-PCR and real-time PCR
Total RNA was extracted from the cells with different treatments by Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturers protocol. Subsequently, the extracted RNA was reverse-transcribed using PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan). The cDNAs were then subjected to SYBR Green-based real-time PCR analysis. The primers used in qRT-PCR assays were listed in Table S1.
2.8. Western blotting
Total protein was extracted with lysis buffer containing protease inhibitors and subjected to quantitative determination. The proteins were separated on SDS-PAGE gels and then transferred onto PVDF membranes (Millipore, Bedford, MA). Subsequently, the membranes were blocked with 5% nonfat dry milk and incubated with specific primary antibodies. After overnight incubation at 4 °C, immunoblots were probed with ECL detection reagent (Millipore) according to standard protocols. The detailed information of primary antibodies was shown in Table S2.
2.9. Chromatin immunoprecipitation (ChIP) assay
For ChIP assay, the SimpleChIP® Enzymatic Chromatin IP Kit (Cell Signaling, Danvers, MA, USA) was used according to the manufacturer's protocol. The precipitated DNA samples were detected with PCR method. The primes used in this experiment were listed in Table S1.
2.10. Immunoprecipitation (IP) assay
The IP/co-IP analysis was performed using a PierceTM Co-Immunoprecipitation Kit (Thermo-Fisher, Waltham, MA, USA) according to the manufacturer's instructions. Protein extracts were incubated with 5 μg antibody.
2.11. Tumor xenograft model
For xenograft model, 3 × 105 BGC-823 cells transfected with CHAF1A or negative control were subcutaneously injected into the right or left flank of 5 thymus-null BALB/c nude (Mu Tu Biological Technology, Nanjing, China), respectively. Tumor growth was monitored every 2 days, for a total period of 16 days. All animal procedures were approved by Shandong University Research Ethics Committee.
2.12. Luciferase reporter assay
Top-Flash constructs (Addgene plasmids 16558), Fop-Flash constructs (Addgene plasmids 16559), CHAF1A promoter wild or binding site mutant plasmids and the internal control vector pRL-TK were transfected into GC cells using Roche Transfection Reagent (Roche, Basel, Switzerland) following the manufacturer's instructions. After 24 h of incubation, luciferase activity were measured using a Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer's protocol.
2.13. Statistical analysis
All experiments were repeated at least three times. Data were presented as mean ± standard deviation. Student t-test or Mann–Whitney U test was used for comparing means between two groups. The difference between the two groups in cell growth curve was determined by repeated measures analysis of variance. For clinical data, statistical analysis was performed using the SPSS version 23.0. Correlation analysis of mRNA data was performed using linear regression. P values of <0.05 were considered statistically significant.
3. Results
3.1. Increased CHAF1A expression predicts poor clinical outcome in GC patients
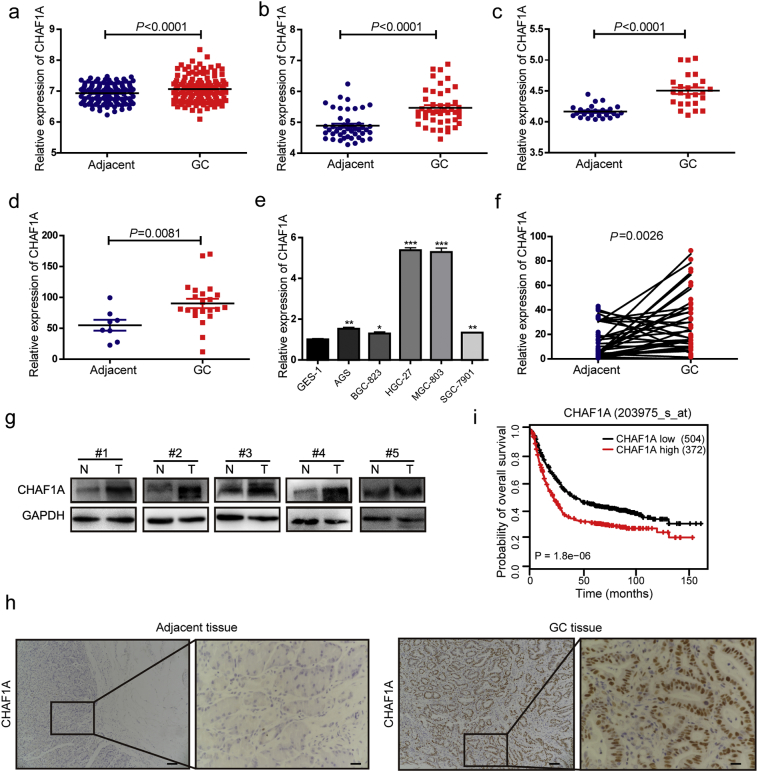
Bioinformatic analysis has been used to discover previously unknown functions of multifarious genes associated with cancer [34,35]. To investigate the potential role of CHAF1A in human GC pathogenesis, we firstly carried out an analysis of GEO databases (e.g. GSE27342, GSE63089, GSE13195 and GSE2685). We found that the mRNA expression of CHAF1A was upregulated in GC tissues compared to adjacent normal tissues (Fig. 1a–d). CHAF1A was also differentially expressed in several gastric cell lines, including GES-1, AGS, BGC-823, HGC-27, MGC-803 and SGC-7901. Of note, the expression of CHAF1A was higher in poorly differentiated HGC-27 and MGC-803 cell lines compared to immortalize epithelial GES-1 cells (Fig. 1e, Supplementary Fig. 1). Subsequently, the results from GEO database analysis were validated in 36 GC biopsies and their corresponding adjacent non-tumor tissues. Indeed, CHAF1A mRNA expression was significantly higher in GC biopsies than in adjacent normal tissues (Fig. 1f). Similarly, CHAF1A protein expression was higher in five GC samples than in adjacent non-tumor tissues (Fig. 1g). Consistent with the above-mentioned findings, immunohistochemistry (IHC) staining revealed the overexpression of CHAF1A in GC patients (Fig. 1h).
Fig. 1.
CHAF1A expression is elevated in GC and associated with poor clinical outcome. a–d GEO RNA sequencing database analysis of the relatively differential expression level of CHAF1A in human GC and paired adjacent normal tissues. The data were derived from the GSE29272 (a), GSE63089 (b), GSE13195 (c) and GSE2685 (d). e Relative expression levels of CHAF1A in different gastric cancer cell lines and immortalized gastric cells GES-1. *p < 0.0 5, **p < 0.01, ***p < 0.001 by Student's t-test. f The mRNA levels of CHAF1A in thirty six pairs of GC and their paired adjacent normal tissues were measured by real-time PCR (n = 36). g The protein levels of CHAF1A in five pairs of GC tissues (T) and adjacent non-tumor tissues (N) measured by western blot. h IHC staining for CHAF1A was performed on GC and adjacent normal tissues. Scale bars: 200 μm (insets 50 μm). i Kaplan-Meier analysis was performed, higher expression of CHAF1A correlated with significantly worse overall survival of GC patients (P = 1.8e-06; n = 876). The data and p-values were obtained from the Kaplan-Meier Plotter database.
In order to determine whether CHAF1A expression is correlated with GC prognosis, Kaplan Meier plotter database was used to assess the effects of CHAF1A on the survival of 876 GC patients. The results showed that CHAF1A overexpression was significantly correlated with poor overall survival of GC patients (Fig. 1i). Taken together, our data imply the oncogenic role of CHAF1A in GC.
3.2. CHAF1A promotes GC cell proliferation in vitro and in vivo
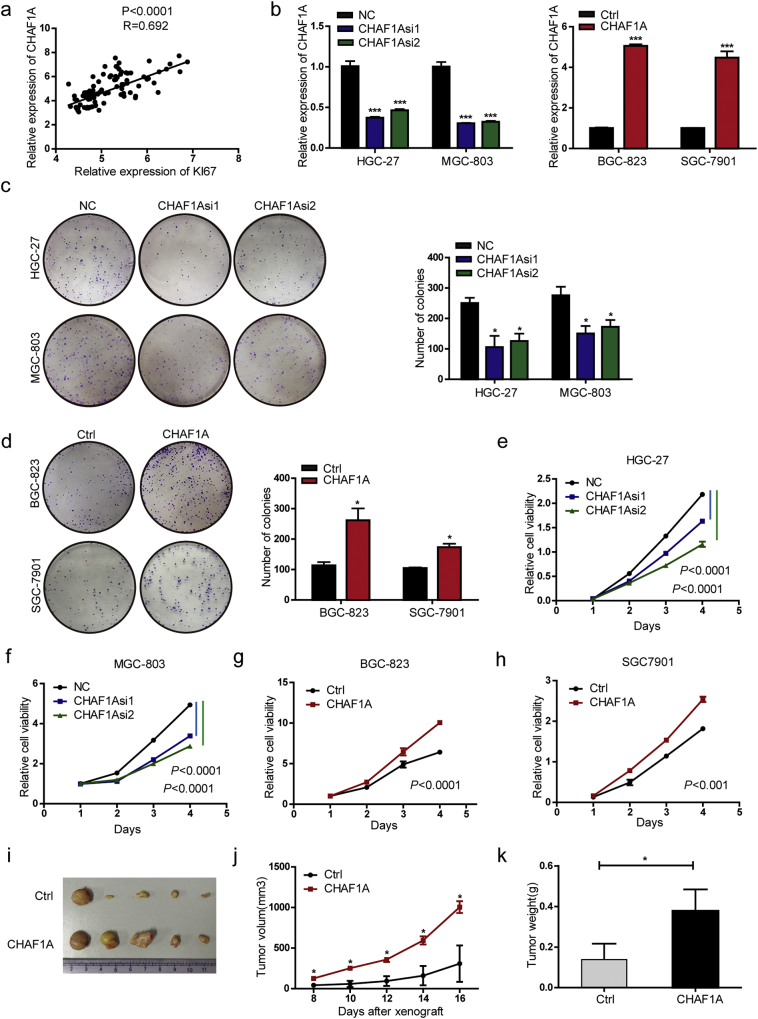
In the next step, the effect of CHAF1A on GC cell proliferation was explored. CHAF1A expression was positively correlated with the proliferation marker Ki67 in 45 pairs of matched GC and normal tissues from GEO datasets (GEO63089), suggesting the potential role of CHAF1A in GC cell proliferation (Fig. 2a). To further evaluate these findings, CHAF1A expression was knocked down in HGC-27 and MGC-803 cells, and overexpressed in BGC-823 and SGC-7901 cells (Fig. 2b). CHAF1A knockdown reduced the colony formation in GC cells, while CHAF1A overexpression exhibited the opposite effects (Fig. 2c–d). Likewise, the same results were obtained for cell viability (Fig. 2e–h).
Fig. 2.
CHAF1A promotes cell proliferation of GC. a Correlation of Ki-67 with CHAF1A mRNA expression in 45 pairs of GC and adjacent non-tumor tissues. The data were derived from the GEO database (GSE63089). b CHAF1A mRNA levels in indicated cells transfected with CHAF1A siRNA or CHAF1A over-expression plasmid. c Representative images (left) and quantification (right) of Negative Control (NC) – or CHAF1A siRNA-transfected HGC-27 and MGC-803 cells analyzed in a colony formation assay. d Representative images (left) and quantification (right) of vector- or CHAF1A plasmid-transfected BGC-823 and SGC-7901 cells analyzed in a colony formation assay. e-f The effect of CHAF1A knockdown on cell viability determined by CCK8 assay in HGC-27 and MGC-803 cells. g-h The effect of CHAF1A overexpression on cell viability determined by CCK8 assay in BGC-823 and SGC-7901 cells. Similar results were repeated in three independent experiments. i-k The effect ectopic CHAF1A expression on tumor formation ability in nude mice xenograft model (i), tumor growth curve (j) and tumor weight (k) in each group. *p < 0.0 5, **p < 0.01, ***p < 0.001 by Student's t-test.
To confirm these findings in vitro, BGC-823 cells with stable CHAF1A overexpression were constructed and injected into nude mice. Gastric tumors formed in CHAF1A overexpression group were larger than those in the control group, with regard to size and weight (Fig. 2i–k). Therefore, our data suggest that CHAF1A can promote gastric cell proliferation in vitro and tumor growth in vivo, indicating its pro-tumor role in GC.
3.3. CHAF1A upregulates the expression of c-MYC and CCND1 in GC
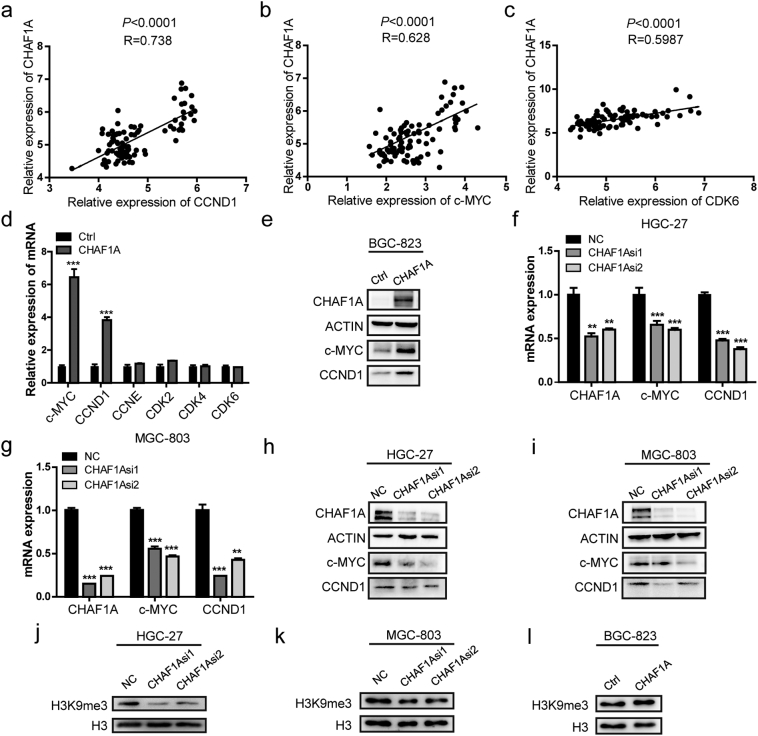
Further, we sought to investigate the molecular mechanism underlying CHAF1A-mediated tumor growth. The expression levels of CHAF1A and several proliferation-associated genes such as CCND1, CDK2, CDK4, CDK6, CCNE1 and c-MYC from GEO datasets (GEO63089) were analyzed. CHAF1A expression was positively correlated with CCND1, c-MYC and CDK6 expression levels in 45 pairs of matched normal and GC tissues, with the R (coefficient of correlation) values of 0.738, 0.628, and 0.5987, respectively. However, no significant relationship was found on other proliferation-associated genes (Fig. 3a–c). Furthermore, we confirmed that CHAF1A expression was positively correlated with c-MYC and CCND1 in the collected 24 GC biopsies and corresponding adjacent normal gastric tissue (Supplementary Fig. 2). Similarly, we observed that CHAF1A overexpression significantly increased the mRNA and protein expression levels of c-MYC and CCND1 in BGC-823 cells (Fig. 3d–e), but not CDK2, CDK4, CDK6 and CCNE1 (Fig. 3d). Accordingly, depletion of CHAF1A demonstrated the opposite effects in HGC-27 and MGC-803 cells (Fig. 3f–i). Collectively, these results indicate that c-MYC and CCND1 are the target genes of CHAF1A. In addition, as CHAF1A was responsible for trimethylation of histone H3 at position lysine 9, we examined the global change of H3K9me3 level. Silencing of CHAF1A significantly reduced the level of global H3K9me3 in HGC-27 and MGC-803 cells, while CHAF1A overexpression exhibited the opposite effects (Fig. 3j–l).
Fig. 3.
CHAF1A participates in the regulation of c-MYC and CCND1 in GC. a–c Correlation of CCND1, c-MYC, CDK6 with CHAF1A mRNA expression in 45 pairs of GC and adjacent non-tumor tissues. The data were derived from the GEO database (GSE63089). d RT–PCR analysis of CCND1, c-MYC, CDK6, CCNE1, CDK2 and CDK4 mRNA expression in BGC-823 cells transfected with CHAF1A or control plasmid. e Western blot analysis of c-MYC and CCND1 level in BGC-823 cells that transfected with CHAF1A or control plasmid. f–g The mRNA expression of c-MYC and CCND1 in HGC-27 and MGC-803 cells transfected with CHAF1A siRNA or negative control. h–i The protein expression of c-MYC and CCND1 in HGC-27 and MGC-803 cells under CHAF1A knockdown. j–k Western blot analysis of the global H3K9me3 levels in HGC-27 and MGC-803 cells transfected with CHAF1A siRNA or negative control. l Western blot analysis of the global H3K9me3 levels in BGC-823 cells transfected with CHAF1A or control plasmid.
3.4. CHAF1A interacts with TCF4 to promote the expression of c-MYC and CCND1 in GC cells
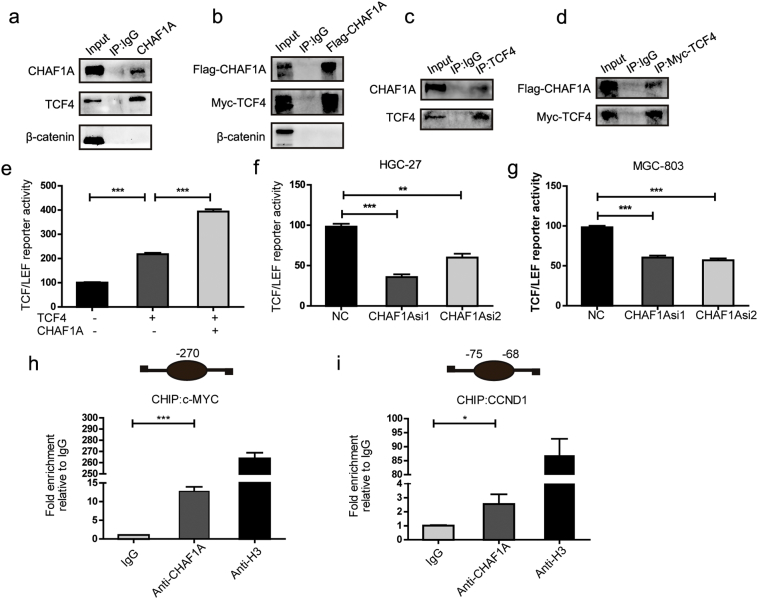
The molecular mechanisms underlying CHAF1A-mediated upregulation of c-MYC and CCND1 were elucidated. Aberrant Wnt signaling pathway plays a crucial role during the development of GC tumors [32,33]. C-MYC and CCND1 are the well-known targets of TCF4/β-catenin transcription factors complex in the activated Wnt signaling pathway [36,37]. Therefore we sought to determine the potential role of CHAF1A in TCF4/β-catenin pathway. First, we examined the effects of CHAF1A depletion on the expression levels of TCF4 and β-catenin in HGC-27 and MGC-803 cells. However, CHAF1A did not affect the mRNA expression of TCF4 and β-catenin in GC cells (Supplementary Fig. 3A). Likewise, the protein levels of β-catenin were not affected by CHAF1A (Supplementary Fig. 3B). Next, we evaluated the endogenous interaction between CHAF1A and Wnt pathway proteins through immunoprecipitation (IP). TCF4 was identified as the protein that directly bound to CHAF1A, whereas no interaction was found between β-catenin and CHAF1A in GC cells (Fig. 4a). The results of CHAF1A-TCF4 interaction were further confirmed by co-immunoprecipitation in GC cells (Fig. 4b). Notably, reciprocal IP and CoIP verified the physical association between CHAF1A and TCF4 (Fig. 4c–d). Then, Wnt-response luciferase reporter assay was conducted to examine the effects of CHAF1A/TCF4 complex on gene expression. The results showed that TCF4 overexpression increased the luciferase activity mediated by Wnt signaling (TOP/FOP), whereas co-transfection of TCF4 and CHAF1A further enhanced their transcriptional activity (Fig. 4e). Consistently, silencing of CHAF1A significantly decreased luciferase activity (Fig. 4f–g). Finally, chromatin immunoprecipitation (ChIP) assays indicated the direct occupancy of CHAF1A on the promoter regions of c-MYC and CCND1 (Fig. 4h–i). Our results also showed that silencing of CHAF1A reduced the binding of TCF4 to the c-MYC and CCND1 promoter regions (Supplementary Fig. 4). Taken altogether, these results suggest that CHAF1A is physically interacted with TCF4 to increase the expression of c-MYC and CCND1 in GC.
Fig. 4.
CHAF1A interacts with TCF4 and promotes transcription of c-MYC and CCND1. a Endogenous CHAF1A directly interacts with endogenous TCF4 rather than β-catenin. CHAF1A was immunoprecipitated with antibodies against CHAF1A and proteins in input, and IP were analyzed by Western blotting using the indicated Abs. b Exogenously expressed Flag-CHAF1A associates with exogenous Myc-tagged TCF4. Flag-CHAF1A was immunoprecipitated with an anti-Flag antibody. c Endogenous TCF4 associates with endogenous CHAF1A. d Exogenously expressed Myc-tagged TCF4 associates with exogenous Flag-CHAF1A followed by co-immunoprecipitation with an anti-Myc antibody. e Regulation of TCF/LEF reporter activity by CHAF1A. BCG-823 cells were transfected with TCF/LEF reporters together with TCF4 vectors alone or TCF4 plus CHAF1A expression vectors and luciferase activity was assessed 48 h post-transfection. f–g The TCF/LEF transcriptional activity in HGC-27 (f) and MGC-803 (g) cells after CHAF1A knockdown. Relative luciferase activities were normalized with the internal control. Quantification are shown here from three independent biological replicates. h–i ChIP assay for CHAF1A occupancy on the promoters of TCF4 targets c-MYC (h) and CCND1 (i). ChIP was performed with chromatin derived from MGC-803 cells. The final DNA samples were amplified by qPCR with pairs of primers as described in Materials and Methods for the TBS in the c-MYC and CCND1 promoter. Histone H3 antibody was used as a positive control. IgG antibody was used as a negative control.
3.5. CHAF1A is overexpressed by Sp1 in GC
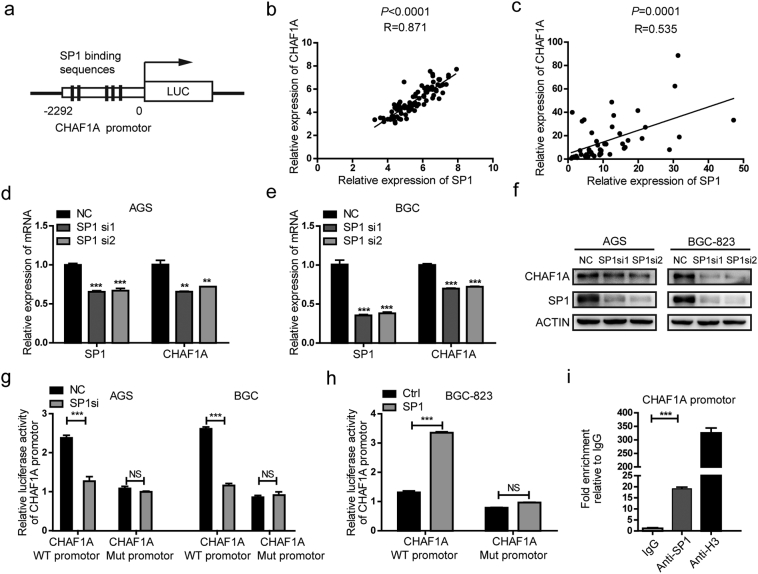
To elucidate the underlying mechanisms of CHAF1A overexpression, the transcription factors binding to specific conservative elements were identified by in silico analysis. The promoter of CHAF1A containing multiple classical GGGCGG elements was recognized by transcriptional factor Sp1, as predicted by PROMO 3.0 (Fig. 5a). Also, CHAF1A expression was positively correlated with Sp1 expression, indicated by GEO dataset (GSE63089) analysis and 24 paired sets of GC biopsies and adjacent normal gastric tissues (Fig. 5b–c). In addition, Sp1 depletion reduced the mRNA and protein expression levels of CHAF1A in GC cell lines (Fig. 5d–f). Knockdown of Sp1 significantly decreased the promoter activity of CHAF1A in GC cells, while Sp1 overexpression exhibited the opposite effects (Fig. 5g–h). Furthermore, ChIP analysis revealed that CHAF1A promoter region was occupied by Sp1 (Fig. 5g). These results highlight that Sp1 is responsible for the overexpression of CHAF1A in GC.
Fig. 5.
CHAF1A was upregulated by Sp1 in GC. a Schematic diagram illustrates the Sp1 binding sites on the promoter region of CHAF1A gene. b–c The positive correlation between CHAF1A and Sp1 in the GEO database (GSE63089) (b) and clinical GC samples (c) using Pearson correlation coefficient analysis. d–e The mRNA expression of CHAF1A in AGS and BCG-823 cells transfected with Sp1 siRNA or negative control. f The protein level of the CHAF1A in AGS and BCG-823 cells under the knockdown of the Sp1. g AGS and BGC-823 cells were transfected with Sp1 siRNA, along with wild-type CHAF1A promoter reporter plasmid or mutant CHAF1A promoter reporter plasmid for 24 h, and then luciferase assay was performed. h The promoter activity of CHAF1A was also performed in BGC-823 cells transfected with Sp1 expression plasmids. i ChIP analysis of Sp1 binds to CHAF1A gene promoter region. Similar results were acquired in three independent experiments. Every bar represents the mean ± s.e.m. of three independent experiments in (d), (e), (g), (h) and (i). **p < 0.01, ***p < 0.001 by Student's t-test.
3.6. CHAF1A expression induced by H. pylori in Sp1 dependent manner
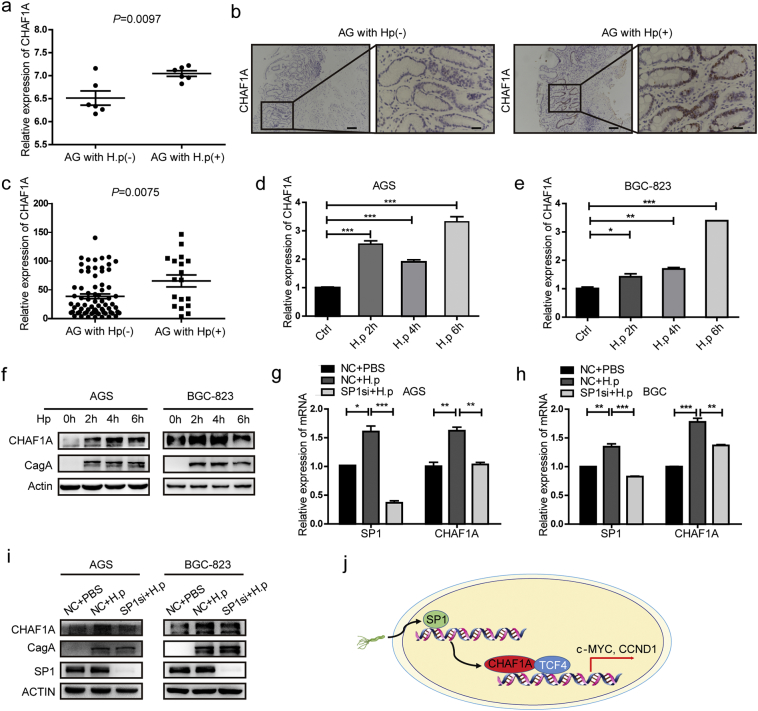
Helicobacter pylori (H. pylori) infection has been associated with an increased risk of gastric adenocarcinoma [38]. We aimed to determine whether H. pylori infection can affect the expression of CHAF1A. The analysis of microarray expression data from GEO dataset (GSE27411) showed that CHAF1A expression was significantly increased in H. pylori-infected gastritis tissues compared to non-infected tissues (Fig. 6a). These findings were validated in the collected H. pylori-positive gastric tissue samples at both mRNA and protein levels (Fig. 6b–c). H. pylori infection significantly increased the mRNA and protein expression levels of CHAF1A in GC cells (Fig. 6d–f). It has been reported that Sp1 can be activated by H. pylori infection [39] and CHAF1A expression was regulated by Sp1 (see the previous data). CHAF1A expression was also positively correlated with Sp1 expression in AG patients (Supplementary Fig. 5). Therefore, we proposed that Sp1 may be associated with H. pylori-mediated CHAF1A expression. To test the hypothesis, Sp1 was depleted in GC cells followed by H. pylori-induced infection. As a consequence, we found that Sp1 depletion abrogated the induction of CHAF1A in H. pylori-infected GC cells (Fig. 6g–i). Collectively, these results indicate that Sp1 may involve in the overexpression of CHAF1A triggered by H. pylori infection.
Fig. 6.
CHAF1A expression induced by H. pylori is Sp1 dependent. a CHAF1A mRNA expression in six pairs of H. pylori-infected and H. pylori non-infected gastric tissue. The data were derived from the GEO database (GSE27411). b IHC staining for CHAF1A was performed on H. pylori non-infected or H. pylori-infected AG samples. Scale bars: 200 μm (insets 50 μm). c The mRNA levels of CHAF1A in H. pylori- infected AG samples (n = 18) and H. pylori non-infected (n = 73). d-e RT–PCR analysis of CHAF1A mRNA in H. pylori-infected AGS (d) and BGC-823 (e) cells at different time points. f The protein expression of CHAF1A in H. pylori -infected AGS and BGC-823 cells at different time points. g–h CHAF1A and Sp1 changes at RNA level with Sp1 siRNA and H. pylori treatment solely or jointly in AGS (g) and BGC-823 (h) cells cells. i Western blot analysis of CHAF1A and Sp1 protein level with Sp1 siRNA and H. pylori treatment solely or jointly in AGS and BGC-823 cells. j Proposed schematic model of oncogenic function of CHAF1A in gastric cancer.
4. Discussion
Increasing evidence has suggested the role of histone chaperones in contributing to tumor progression. For instance, the Anti-Silencing Function 1A (ASF1A) is upregulated in primary gastrointestinal cancer and predicts shorter overall survival in patients with colorectal cancer [40]. The expression of Aprataxin PNK-like factor (APLF) is also enhanced in breast cancer and can regulate metastasis-associated EMT in invasive breast cancer [41]. Moreover, high levels of Facilitates Chromatin Transcription (FACT) complex have been associated with the tumor progression and the poorly differentiated carcinomas [42]. In addition, histone cell cycle regulator (HIRA) has been reported to enhance β-catenin expression by recruiting histone-lysine N-methyltransferase Setd1A [43]. In this study, we focused on CHAF1A and revealed its oncogenic roles in GC development and progression. CHAF1A was overexpressed in GC cell lines and primary tumors. Increased expression of CHAF1A can predict a poor prognosis in patients with GC.
CHAF1A promoted gastric carcinogenesis by upregulating c-MYC and CCND1. To our surprise, we found a direct interaction between TCF4 and CHAF1A, which may account for the enhanced expression of c-MYC and CCND1. However, whether the interaction between TCF4 and CHAF1A is due to the histone chaperone function remains unclear. Therefore, more complex functional studies are warranted in the future to clarify the histone chaperone independent function of CHAF1A. Apart from the Wnt pathway, there are many other pathways, such as NF-kB and PI3K/AKT pathways, that have been implicated in gastric carcinogenesis [44]. Thus, it is essential to investigate the relationship of CHAF1A with these pathways, and to determine their potential involvement in human carcinogenesis.
Indeed, the high expression of CHAF1A was observed in GC. However, the molecular mechanisms of CHAF1A overexpression remain largely unknown. In this study, we investigated the molecular mechanisms underlying CHAF1A upregulation in GC. Interestingly, Sp1 was identified as the factor corresponding to the enhanced transcription of CHAF1A. Transcription factors such as E2F, AP-1 and p53, also serve as promising potential regulators. Other possible factors underlying CHAF1A overexpression may include epigenetic regulation. For instance, DNA methylation, histone modification and microRNA dysregulation can lead to the overexpression of CHAF1A in GC.
Infection with H. pylori is a well-known risk factor for gastric neoplasia [45]. To the best of our knowledge, this is the first study describing that H. pylori can induce the expression of histone chaperone CHAF1A. Even though we discovered that Sp1 can modulate the induction of CHAF1A triggered by H. pylori, the exact mechanisms during the pathogenic processes of GC remained elusive. More studies are needed to elucidate the potential molecular mechanisms, which will help us to understand the immunopathogenesis of gastric disease associated with H. pylori infection.
CHAF1A has also been shown to modulate DNA methylation by forming a complex with MBD1 and SETDB1 [18]. Furthermore, previous studies demonstrated that CHAF1A regulated the H3K9me3 epigenetic marker of heterochromatin domains in pluripotent embryonic cells [46]. In this study, we found similar role of CHAF1A in regulating the global H3K9me3 in GC cells.
In summary, this study identified a novel oncogenic role of CHAF1A in GC. The upregulation of CHAF1A can promote GC cell growth and predict poor prognosis in GC patients. Hence, CHAF1A may serve as a potential target for the prevention and treatment of GC.
Funding sources
This work was supported by the National Natural Science Foundation of China (Nos. 81772151, 81571960, 81871620, 81670146 and 81470318), the Shandong Provincial Major Scientific and Technical Innovation Project (2018CXGC1208) and the training project of Shandong Provincial Natural Science Foundation (ZR2018PH013).
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
JJ designed the study. LZ, SL, TL, WS, LM, WS, XJ and PS performed experiments. LZ, XL, CC and JJ analyzed data. XL, CC and JJ obtained funding. LZ and JJ prepared the figures. LZ, XL and JJ wrote the manuscript. JJ supervised the study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.11.009.
Appendix A. Supplementary data
Supplementary material
References
- 1.Fitzmaurice C., Dicker D., Pain A. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dassen A.E., Dikken J.L., van de Velde C.J. Changes in treatment patterns and their influence on long-term survival in patients with stages I-III gastric cancer in the Netherlands. Int. J. Cancer. 2013;133(8):1859–1866. doi: 10.1002/ijc.28192. [DOI] [PubMed] [Google Scholar]
- 3.Colquhoun A., Arnold M., Ferlay J. Global patterns of cardia and non-cardia gastric cancer incidence in 2012. Gut. 2015;64(12):1881–1888. doi: 10.1136/gutjnl-2014-308915. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B., Papadopoulos N., Velculescu V.E. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarris M., Nikolaou K., Talianidis I. Context-specific regulation of cancer epigenomes by histone and transcription factor methylation. Oncogene. 2014;33(10):1207–1217. doi: 10.1038/onc.2013.87. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Paredes M., Esteller M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 2011;17(3):330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 7.Ozdag H., Teschendorff A.E., Ahmed A.A. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics. 2006;7:90. doi: 10.1186/1471-2164-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng J., Ge Z., Wang L. The histone demethylase RBP2 is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology. 2010;138(3):981–992. doi: 10.1053/j.gastro.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Vardabasso C., Hasson D., Ratnakumar K. Histone variants: emerging players in cancer biology. Cell. Mol. Life Sci. 2014;71(3):379–404. doi: 10.1007/s00018-013-1343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess R.J., Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 2013;20(1):14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verreault A., Kaufman P.D., Kobayashi R. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87(1):95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 12.Takami Y., Ono T., Fukagawa T. Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol. Biol. Cell. 2007;18(1):129–141. doi: 10.1091/mbc.E06-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadyrova L.Y., Blanko E.R., Kadyrov F.A. CAF-I-dependent control of degradation of the discontinuous strands during mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 2011;108(7):2753–2758. doi: 10.1073/pnas.1015914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyen C.M., Moshkin Y.M., Chalkley G.E. Subunits of the histone chaperone CAF1 also mediate assembly of protamine-based chromatin. Cell Rep. 2013;4(1):59–65. doi: 10.1016/j.celrep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Smith C.L., Matheson T.D., Trombly D.J. A separable domain of the p150 subunit of human chromatin assembly factor-1 promotes protein and chromosome associations with nucleoli. Mol. Biol. Cell. 2014;25(18):2866–2881. doi: 10.1091/mbc.E14-05-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tchenio T., Casella J.F., Heidmann T. A truncated form of the human CAF-1 p150 subunit impairs the maintenance of transcriptional gene silencing in mammalian cells. Mol. Cell. Biol. 2001;21(6):1953–1961. doi: 10.1128/MCB.21.6.1953-1961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murzina N., Verreault A., Laue E. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell. 1999;4(4):529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 18.Reese B.E., Bachman K.E., Baylin S.B. The methyl-CpG binding protein MBD1 interacts with the p150 subunit of chromatin assembly factor 1. Mol. Cell. Biol. 2003;23(9):3226–3236. doi: 10.1128/MCB.23.9.3226-3236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarraf S.A., Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004;15(4):595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 20.Heyd F., Chen R., Afshar K. The p150 subunit of the histone chaperone Caf-1 interacts with the transcriptional repressor Gfi1. Biochim. Biophys. Acta. 2011;1809(4–6):255–261. doi: 10.1016/j.bbagrm.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Wu Z., Cui F., Yu F. Up-regulation of CHAF1A, a poor prognostic factor, facilitates cell proliferation of colon cancer. Biochem. Biophys. Res. Commun. 2014;449(2):208–215. doi: 10.1016/j.bbrc.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Glinsky G.V., Glinskii A.B., Stephenson A.J. Gene expression profiling predicts clinical outcome of prostate cancer. J. Clin. Invest. 2004;113(6):913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M., Jia Y., Liu Z. Chromatin assembly factor 1, subunit A (P150) facilitates cell proliferation in human hepatocellular carcinoma. Onco. Targets Ther. 2016;9:4023–4035. doi: 10.2147/OTT.S107050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bethke L., Webb E., Murray A. Comprehensive analysis of the role of DNA repair gene polymorphisms on risk of glioma. Hum. Mol. Genet. 2008;17(6):800–805. doi: 10.1093/hmg/ddm351. [DOI] [PubMed] [Google Scholar]
- 25.Peng H., Du B., Jiang H. Over-expression of CHAF1A promotes cell proliferation and apoptosis resistance in glioblastoma cells via AKT/FOXO3a/Bim pathway. Biochem. Biophys. Res. Commun. 2016;469(4):1111–1116. doi: 10.1016/j.bbrc.2015.12.111. [DOI] [PubMed] [Google Scholar]
- 26.Barbieri E., De Preter K., Capasso M. Histone chaperone CHAF1A inhibits differentiation and promotes aggressive neuroblastoma. Cancer Res. 2014;74(3):765–774. doi: 10.1158/0008-5472.CAN-13-1315. [DOI] [PubMed] [Google Scholar]
- 27.Barbieri E., De Preter K., Capasso M. A p53 drug response signature identifies prognostic genes in high-risk neuroblastoma. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0079843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Clevers H., Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 31.Macdonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ooi C.H., Ivanova T., Wu J. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5(10) doi: 10.1371/journal.pgen.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao J., Fan S., Ma W. Roles of Wnt/beta-catenin signaling in the gastric cancer stem cells proliferation and salinomycin treatment. Cell Death Dis. 2014;5:e1039. doi: 10.1038/cddis.2013.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park Y.Y., Jung S.Y., Jennings N.B. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis. 2012;33(10):1843–1853. doi: 10.1093/carcin/bgs167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang M.H., Choi H., Oshima M. Estrogen-related receptor gamma functions as a tumor suppressor in gastric cancer. Nat. Commun. 2018;9(1):1920. doi: 10.1038/s41467-018-04244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tetsu O., Mccormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398(6726):422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 37.He T.C., Sparks A.B., Rago C. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 38.Wadhwa R., Song S., Lee J.S. Gastric cancer-molecular and clinical dimensions. Nat. Rev. Clin. Oncol. 2013;10(11):643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strowski M.Z., Cramer T., Schafer G. Helicobacter pylori stimulates host vascular endothelial growth factor-A (vegf-A) gene expression via MEK/ERK-dependent activation of Sp1 and Sp3. FASEB J. 2004;18(1):218–220. doi: 10.1096/fj.03-0055fje. [DOI] [PubMed] [Google Scholar]
- 40.Liang X., Yuan X., Yu J. Histone Chaperone ASF1A predicts poor outcomes for patients with Gastrointestinal Cancer and Drives Cancer Progression by Stimulating Transcription of beta-Catenin Target Genes. EBioMedicine. 2017;21:104–116. doi: 10.1016/j.ebiom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majumder A., Syed K.M., Mukherjee A. Enhanced expression of histone chaperone APLF associate with breast cancer. Mol. Cancer. 2018;17(1):76. doi: 10.1186/s12943-018-0826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia H., Miecznikowski J.C., Safina A. Facilitates chromatin transcription complex is an "accelerator" of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 2013;4(1):159–173. doi: 10.1016/j.celrep.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Jiao J. Histone chaperone HIRA regulates neural progenitor cell proliferation and neurogenesis via beta-catenin. J. Cell Biol. 2017;216(7):1975–1992. doi: 10.1083/jcb.201610014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye B., Jiang L.L., Xu H.T. Expression of PI3K/AKT pathway in gastric cancer and its blockade suppresses tumor growth and metastasis. Int. J. Immunopathol. Pharmacol. 2012;25(3):627–636. doi: 10.1177/039463201202500309. [DOI] [PubMed] [Google Scholar]
- 45.Hartgrink H.H., Jansen E.P., van Grieken N.C. Gastric cancer. Lancet. 2009;374(9688):477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houlard M., Berlivet S., Probst A.V. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material