FIGURE 4.
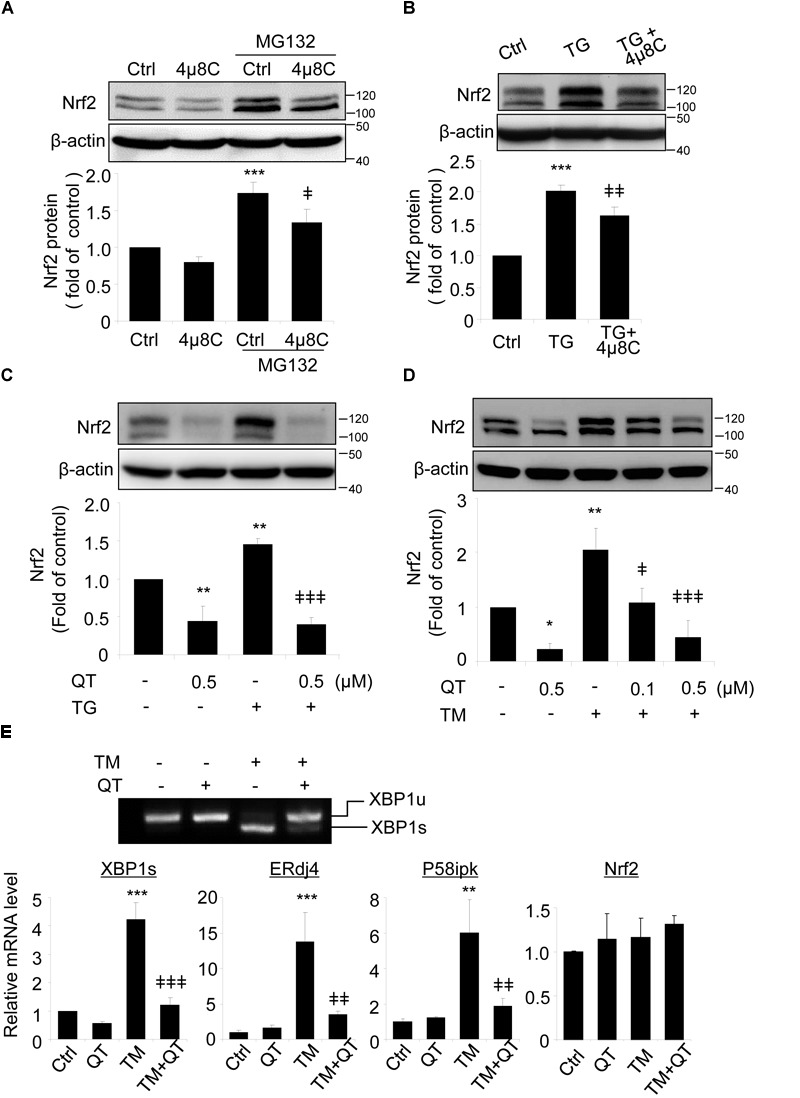
XBP1 splicing inhibitors reduce Nrf2 expression in ARPE-19 cells. (A) ARPE-19 cells were treated with proteasome inhibitor MG132 (10 μM, 20 min) with or without pre-treatment of 4μ8C, an XBP1 splicing inhibitor (25 μM, 8 h). (B) ARPE-19 cells were treated with ER stress inducer thapsigargin (TG) (10 μM) with or without 4μ8C (25 μM) for 8 h. Nrf2 expression was detected by Western blot and semi-quantified by densitometry (mean ± SD, n = 3, ∗∗∗p < 0.001 vs. control, ‡p < 0.05, ‡‡p < 0.01 vs. MG132 or TG treated cells). (C,D) ARPE-19 cells were treated with ER stress inducer thapsigargin (TG) (10 μM) or tunicamycin (TM) (50 ng/ml) with or without quinotrierixin (QT) for 8 h, Nrf2 expression was detected by Western blot and semi-quantified by densitometry. (E) ARPE-19 cells were treated with TM (50 ng/ml) with or without QT (0.5 μM) for 8 h, XBP1 splicing was detected by RT-PCR. The levels of spliced XBP1, ERdj4, P58ipk and Nrf2 mRNA were determined by real-time PCR. (mean ± SD, n = 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 vs. control, ‡p < 0.05, ‡‡p < 0.01, ‡‡‡p < 0.001 vs. TM or TG treated cells).
