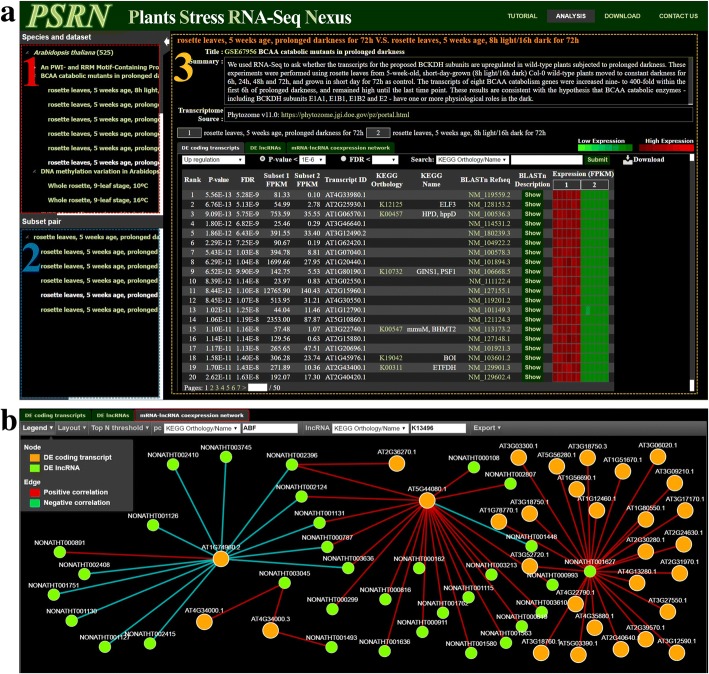
Fig. 2.
Screenshots of the web interface of PSRN. (a) The PSRN user interface consists of three major panels as follows: (1) Species and subset panel: users can browse plant species and phenotype-specific subsets. (2) Subset-pair panel: given the phenotype-specific subset, users can select an associated subset pair. (3) Expression profile panel: subset-pair information and differentially expressed protein-coding transcripts are shown in the panel and sorted by significance level. When users use Search in the expression panel, they can input a transcript ID, KEGG Orthology/name, or RefSeq ID into the autocomplete field that allows for quickly searching and selecting the partially matched terms. Furthermore, by entering more characters, it will filter down the list to better matches. At last, PSRN generates the expression profiles of all isoforms in the search results. In addition, users can download the result generated from PSRN as a file with PDF format. Moreover, users can select a P value or FDR threshold and regulation type to identify significantly differentially expressed transcripts. If a user clicks “DE lncRNAs” in Arabidopsis thaliana and Oryza sativa, differentially expressed lncRNA transcripts are replaced with protein-coding isoforms. (b) The lncRNA regulatory network function in A. thaliana and O. sativa presents the regulatory network according to the correlation of expression between lncRNAs and protein-coding transcripts