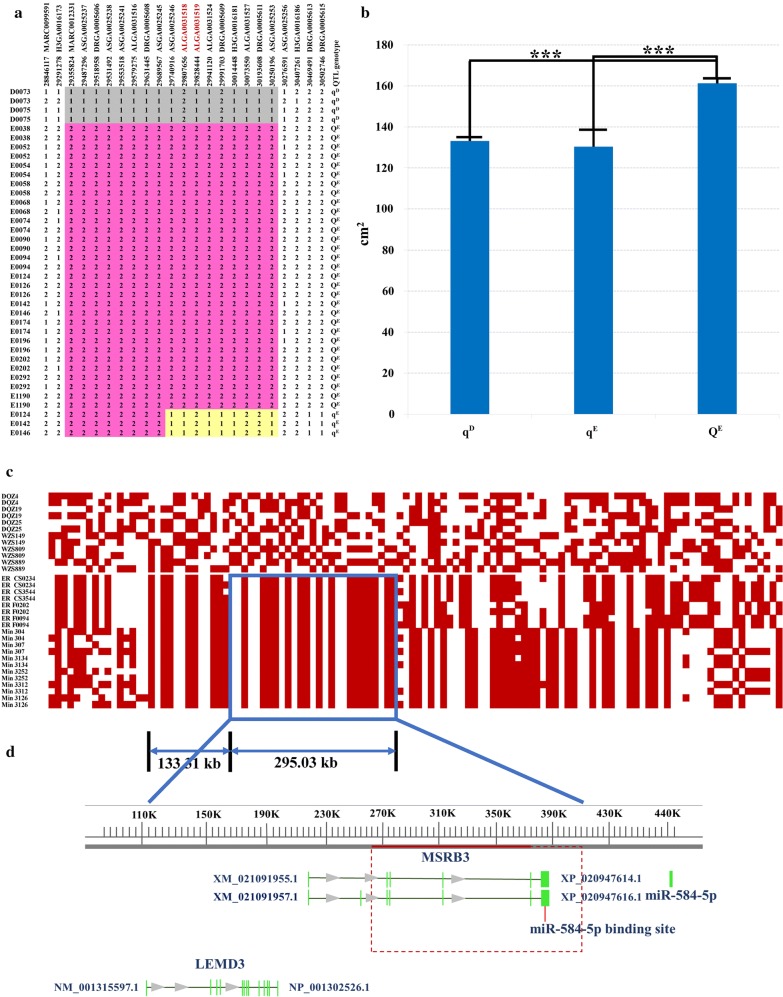
Fig. 3.
Haplotype-sharing analysis refines the critical QTL region to a 137.85 kb interval. a Haplotype-sharing analysis in the 19 F0 founder animals of the White Duroc × Erhualian F2 resource population. The SNPs and their positions are displayed at the top of the figure. For SNPs, alleles with the highest frequencies are denoted 1, while those with the lowest frequencies are denoted 2. The shared haplotypes are indicated by shaded boxes. Identities of these F0 animals are given on the left axis. D and E represent Duroc and Erhualian, respectively. The QTL genotype for each chromosome is shown on the right axis. The red colored box represents the shared Q-haplotype block in Erhualian sows, Individuals E0124, E0142 and E0146 carried a distinct haplotype that is marked in yellow. b Effects of haplotypes qD, QE and qE on ear size in the White Duroc × Erhualian intercross (least square mean ± s.e., cm2). The haplotype QE significantly increased ear size. c Haplotype-sharing analysis in pigs with different ear sizes. The upper panel shows haplotypes of small-eared Tibetan and Wuzhishan pigs that were selected as controls. The lower panel depicts haplotypes of Min and Erhualian pigs with large and floppy ears. The sample ID is shown in the left column and the SNP ID is shown in the top row. The Min and Erhualian pigs shared two haplotypes of 133.31 and 295.03 kb, which are not found in small-eared pigs. d The most likely critical interval of this QTL was defined as the 137.85-kb region indicated by a red box. Exons 3–7 of MSRB3 are located in this region. The gray arrows indicate the direction of the transcripts and the green vertical bars show the exons of the transcripts