Abstract
Long non-coding RNAs (lncRNAs) have critical roles in various types of cancer, but their roles in the development of melanoma and the underlying molecular mechanisms remain to be fully elucidated. In the present study, the role of zinc finger E-box binding homeobox 1 antisense RNA 1 (ZEB1-AS1) in melanoma was assessed. The expression levels of ZEB1-AS1 were increased in melanoma cell lines and tumor tissues as indicated by reverse transcription-quantitative polymerase chain reaction analysis. Kaplan-Meier analysis suggested that higher expression of ZEB1-AS1 predicts poor prognosis of melanoma patients. Furthermore, knockdown of ZEB1-AS1 inhibited the proliferation, migration and invasion of melanoma, suggesting a role of ZEB1-AS1 in the development and progression of melanoma. In addition, a luciferase reporter assay confirmed that the expression of miR-1224-5p was directly regulated by ZEB1-AS1. Transfection with miR-1224-5p mimics reduced the levels of ZEB1-AS1 and the proliferation, migration and invasion of melanoma. In conclusion, ZEB1-AS1 enhances the proliferation, migration and invasion of melanoma, at least in part by inhibiting the expression of miR-1224-5p, and its overexpression is associated with poor survival of melanoma patients. In addition, the ZEB1-AS1/miR-1224-5p interaction may be a promising therapeutic target for melanoma treatment.
Keywords: ZEB1-AS1, microRNA-1224-5p, melanoma, proliferation, invasion
Introduction
Melanoma is a type of malignant tumor derived from melanocytes, which are abundant in the skin and are also present in the mucosa, intestines and eye (1). In 2015, an estimated 3.1 million cases of melanoma were diagnosed globally, and 59,800 associated mortalities were registered (2). Melanoma is the most malignant tumor type in the skin with a high possibility of distant metastasis (3). At present, skin biopsy is widely used for clinical diagnosis. This is usually followed by an extensive excision of the affected scar or tumor (4). However, the 5-year survival rates of patients with melanoma at stage I, II, III and IV are only 94, 44, 38 and 4.6%, respectively (5). Therefore, it is urgent to further investigate the molecular mechanisms of melanoma in order to develop novel strategies for its early diagnosis and treatment.
Long non-coding RNAs (lncRNAs) are transcripts of >200 nucleotides in length that are not translated into protein (6). An increasing number of studies confirmed that certain lncRNAs participate in carcinogenesis and metastasis of melanoma. For instance, long intergenic non-protein coding RNA 673 has been reported to increase melanoma cell invasion by upregulating the expression of matrix metalloproteinase 9, and further affects melanoma survival (7). Furthermore, Wei et al (8) confirmed that urothelial cancer associated 1 (UCA1) was negatively correlated with microRNA (miR)-507, while it was positively correlated with forkhead box (FOX)M1, and that the UCA1/miR-507/FOXM1 axis participates in the invasion, proliferation and changes in the cell cycle associated with the pathogenesis of melanoma. In vivo, metastasis-associated lung adenocarcinoma transcript 1 was also identified as a critical regulator in melanoma development by participating in integrin subunit β1 signal activation (9).
lncRNA zinc finger E-box binding homeobox 1 (ZEB1) antisense RNA 1 (ZEB1-AS1) originates from the promoters of ZEB1. The molecular mechanisms of action of ZEB1-AS1 have been investigated in various diseases, including osteosarcoma (10), glioma (11), prostate cancer (12) and non-small cell lung carcinoma (13). In a previous study, ZEB1-AS1 was confirmed to promote metastasis of hepatocellular tumors and it was indicated to serve as a biomarker to predict poor prognosis (14). Furthermore, ZEB1-AS1 was also confirmed to regulate the level of miR-200s and then participate in cell proliferation and migration of osteosarcoma (15).
While its role in the pathogenesis of cancer has been previously studied, the molecular mechanisms of the roles of ZEB1-AS1 in melanoma have remained largely elusive. Therefore, the detailed mechanisms of action of ZEB1-AS1 and its target miRNAs were herein investigated. The present study may provide novel biomarkers or targets for the diagnosis and treatment of melanoma.
Materials and methods
Sample collection
Melanoma and adjacent normal tissues were collected from 46 melanoma patients (24 males, 22 females; age, 54±13 years) at Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, (Xiangyang, China) between February 2013 to April 2016. All tissue samples were obtained and then frozen and stored in liquid nitrogen at −80°C. According to the median value of ZEB1-AS1, these samples were divided into a ZEB1-AS1 high expression group and low expression group. The present study was approved by the ethics committee of Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, (Xiangyang, China) and written informed consent was obtained by all patients.
Cell lines and culture
A total of 4 human melanoma cell lines, namely SK-MEL-2, WM35, A375 and SK-MEL-5, were obtained from the Cell Bank of The Chinese Academy of Sciences (Shanghai, China). Human primary normal epidermal melanocytes (HEMa; cat. no. PCS-200-013™) were obtained from the American Type Culture Collection (Manassas, VA, USA). All of the cell lines were cultured in RPMI-1640 medium supplemented with fetal bovine serum (10% v/v), penicillin (50 U/ml) and streptomycin (50 U/ml) (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and incubated in a humidified incubator (37°C and 5% CO2).
Transfection
Specific small interfering (si)RNAs against ZEB1-AS1 (siZEB1-AS1#1, 5′-GGAGGUGACCUACAGUUGAAG-3′; and siZEB1-AS1#2, 5′-CGAGAGACCCUGUCUCAAAUA-3′), the scrambled negative control (siNC, 5′-AAUUCUCCGAACGUGUCACGU-3′), miR-1224-5p mimics (5′-GUGAGGACUCGGGAGGUGG-3′) and miR-NC (5′-ACAUCUGCGUAAGAUUCGAGUCUA-3′) were synthesized by Wuhan Genesil Biotechnology Co., Ltd (Wuhan, China). For transfection, Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used according to the manufacturer's protocol (16).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted and purified from cells, melanoma tissues and adjacent normal tissues by using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to manufacturer's protocol and as described previously (17). The purity of the total RNA was confirmed by measuring its absorbance ratio at 260 nm vs. 280 nm using Nanodrop2000 spectrophotometry (Thermo Fisher Scientific, Inc.). Total RNA (1 µg) was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) with gDNA Eraser (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. qPCR was performed using SYBR select master mix (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The thermocycling conditions were as follows: Denaturation at 95°C for 10 min; followed by 40 cycles of denaturation at 95°C for 15 sec and elongation at 60°C for 1 min. The reaction was performed in an ABI 7500 system (Thermo Fisher Scientific, Inc.) and the gene expression was calculated using the 2−ΔΔCq method (18). Small nuclear RNA U6 was used as the reference. The primer sequences were as follows: U6 forward, 5′-AACGAGACGACGACAGAC-3′ and reverse, 5′-GCAAATTCGTGAAGCGTTCCATA-3′; miR-1224-5p forward, 5′-AACGAGACGACGACAGAC-3′ and reverse, 5′-GTGAGGACTCGGGAGGTGG-3′; ZEB1-AS1 forward, 5′-GAGAACGTGGTGGAATCAGA-3′ and reverse, 5′-TCCCATCCTCTTTCTTGTCC-3′.
Cell proliferation assay
A Cell Counting Kit-8 (CCK8) was purchased from Dojindo Laboratories (Kumamoto, Japan) and used to determine the cell proliferation. Based on the manufacturer's protocol, the transfected cells were seeded into 96-well plates at a density of 2×103 cells/well and incubated at 37°C for set time-points (24–96 h, every 24 h). Then cells were incubated with CCK8 at 37°C for 1 h (19). The absorbance was detected by a multi-detection microplate reader (Tecan Group, Ltd, Maennedorf, Switzerland).
Cell invasion and migration assay
For each transfection group, cells were transfected for 48 h. A serum-free cell suspension was then prepared, which was seeded into the upper wells of Transwell plates (8 µm pore; Corning, Incorporated, Corning, NY, USA) with a cell density of 2×104 cells/well. RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum was added to the lower chamber. Subsequently, the plate was incubated at 37°C for 24 h. The cells that had transgressed to the lower side of the membrane were then fixed with 4% formaldehyde for 5 min at 25°C, stained with 0.1% crystal violet for 10 min at 25°C, and images of 5 randomly selected fields of view were captured under an inverted microscope. For the invasive ability assay, the Matrigel® (1:10 dilution in 100 µl; Corning, Incorporated) was evenly spread in the upper chambers of the Transwell insert, followed by solidification in an incubator at 37°C for 1 h. The remaining steps were identical to those for the migration assay.
Luciferase reporter assay
By using a bioinformatics analysis (miRDB tool; http://mirdb.org/miRDB/index.html), miR-1224-5p was identified as a putative target of ZEB1-AS1. The wild-type (WT) and mutant-type (Mut) miR-1224-5p binding sequences of ZEB1-AS1 (synthesized by Sangon Biotech Co., Ltd., Shanghai, China) were respectively inserted into the luciferase reporter vector pmirGLO (Promega Corp., Madison, WI, USA). For the luciferase reporter assay, miR-NC or miR-1224-5p mimics and reporter plasmid were co-transfected into 2×104 A375 cells using Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After transfection for 48 h, the luciferase activity of the sample was detected using the Dual-Luciferase Reporter Assay System (E1910; Promega Corp.) according to the manufacturer's protocols with normalization to the luciferase activity of Renilla.
Statistical analysis
Values are expressed as the mean ± standard deviation. SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. Student's t-test (for two-group comparisons) and one-way analysis of variance followed by Tukey's post-hoc test (for multiple-group comparisons) were performed. The association between clinicopathological features and ZEB1-AS1 expression was analyzed using the Chi-squared test. The association between ZEB1-AS1 levels and overall survival was estimated using the Kaplan-Meier method, and differences between groups were evaluated using the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
ZEB1-AS1 is increased in melanoma
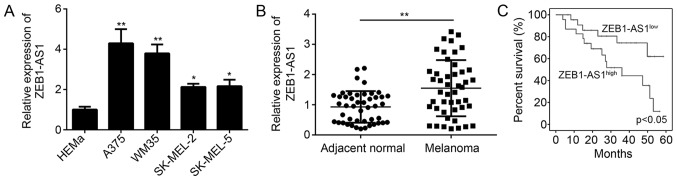
In the present study, ZEB1-AS1 expression levels were determined in melanoma cells and tissues by using RT-qPCR. The results confirmed that ZEB1-AS1 was significantly upregulated in melanoma cell lines, including A375, WM35, SK-MEL-2 and SK-MEL-5 cells, compared with that in normal HEMa cells (Fig. 1A). The relative expression levels of ZEB1-AS1 were highest in the A375 cell line (Fig. 1A). Compared with the expression in adjacent normal tissues, ZEB1-AS1 was significantly upregulated in melanoma tissues (P<0.05; Fig. 1B). In addition, Kaplan-Meier analysis confirmed that higher levels of ZEB1-AS1 (The median value of ZEB1-AS1 was used as cutoff to divide samples into two subgroups) expression predict poor overall survival of patients with melanoma (Fig. 1C). These results suggested that ZEB1-AS1 was upregulated in melanoma tissues and that the expression level is significantly associated with overall survival. The association between clinicopathological characteristics and ZEB1-AS1 expression in malignant melanoma tissues is presented in Table I. The high expression of ZEB1-AS1 was positively associated with advanced TNM stage and lymph node metastasis (Table I), indicating that ZEB1-AS1 may promote the progression of melanoma.
Figure 1.
Expression of ZEB1-AS1 in melanoma and its effect on overall survival. (A) Reverse transcription-quanitative polymerase chain reaction analysis of various melanoma cell lines and HEMa cells indicated that ZEB1-AS1 is overexpressed in melanoma cells. (B) Compared to normal tissue (n=46), ZEB1-AS1 is highly expressed in melanoma tissue (n=46). (C) Kaplan-Meier analysis of overall survival of melanoma patients stratified based on ZEB1-AS1 expression (high or low). Higher levels of ZEB1-AS1 expression predict poor overall survival. *P<0.05, **P<0.01 vs. normal/HEMa. ZEB1-AS1, zinc finger E-box binding homeobox 1 antisense RNA 1; HEMa, human primary normal epidermal melanocytes.
Table I.
Association between clinicopathological features and zinc finger E-box binding homeobox 1 antisense RNA 1 expression (low vs. high) in malignant melanoma tissues.
| Feature | Low (n=23) | High (n=23) | P-value |
|---|---|---|---|
| Age (years) | 0.189 | ||
| ≤60 | 14 | 19 | |
| >60 | 9 | 4 | |
| Sex | 0.768 | ||
| Male | 13 | 11 | |
| Female | 10 | 12 | |
| TNM stage | 0.037 | ||
| I+II | 16 | 8 | |
| III+IV | 7 | 15 | |
| Lymph node metastasis | 0.047 | ||
| No | 15 | 5 | |
| Yes | 8 | 18 |
TNM, tumor-nodes-metastasis.
Knockdown of ZEB1-AS1 inhibits melanoma cell proliferation, invasion and migration
To assess the roles of ZEB1-AS1 in melanoma, specific siRNAs targeting ZEB1-AS1 (siZEB1-AS1#1 and siZEB1-AS1#2) and siNC were individually transfected into the A375 cell line, and the knockdown efficiency was confirmed by RT-qPCR (Fig. 2A). The CCK8 assay was then performed to assess the effect of ZEB1-AS1 knockdown on melanoma cell proliferation. Compared with that in the siNC group, knockdown of ZEB1-AS1 significantly decreased the viability of A375 cells (Fig. 2B), indicating that ZEB1-AS1 has a role in melanoma cell proliferation. Furthermore, Transwell assays suggested that knockdown of ZEB1-AS1 significantly inhibited the migration and invasion of A375 cells (P<0.05; Fig. 2C and D). These results demonstrate that ZEB1-AS1 promotes the malignant behavior of melanoma cells.
Figure 2.
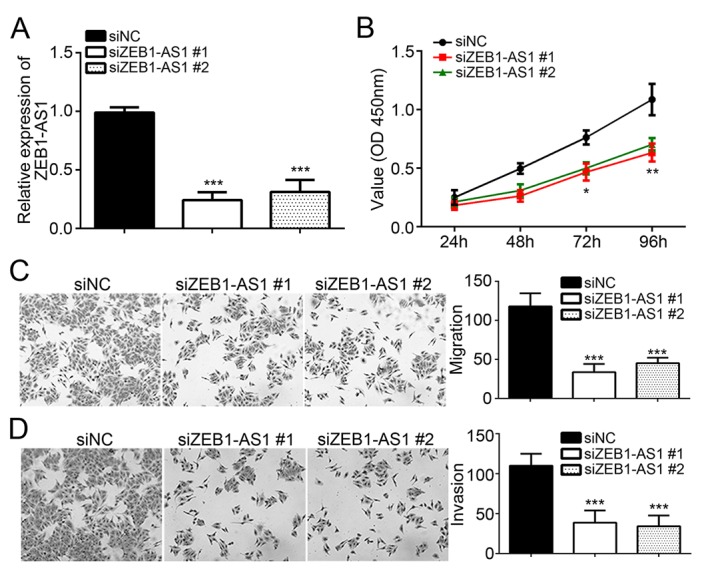
Knockdown of ZEB1-AS1 suppresses melanoma cell proliferation, migration and invasion. (A) ZEB1-AS1 expression levels were significantly decreased in A375 cells transfected with siZEB1-AS1. (B) A Cell Counting Kit-8 assay indicated that knockdown of ZEB1-AS1 inhibited cell proliferation. (C and D) Transwell assays demonstrated that knockdown of ZEB1-AS1 significantly inhibited the migration and invasion of A375 cells (magnification, ×100). *P<0.05, **P<0.01, ***P<0.001 vs. siNC. ZEB1-AS1, zinc finger E-box binding homeobox 1 antisense RNA 1; siNC, scrambled control small interfering RNA; siZEB1-AS1, small interfering RNA targeting ZEB1-AS1; OD, optical density.
miR-1224-5p is directly regulated by ZEB1-AS1
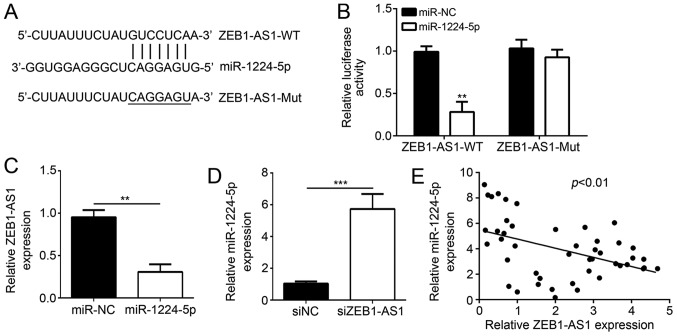
A search with a bioinformatics tool indicated that miR-1224-5p is a target of ZEB1-AS1 and the corresponding binding sites were determined (Fig. 3A). A luciferase activity assay was then employed to verify this interaction in vitro. A375 cells were co-transfected with miR-1224-5p mimics and ZEB1-AS1-WT or ZEB1-AS1-Mut recombinant luciferase reporter plasmids. The results indicated that miR-1224-5p mimics had no significant effect on the luciferase activity in the mutant ZEB1-AS1-Mut/miR-1224-5p plasmid group; however, the luciferase activity in the ZEB1-AS1-WT/miR-1224-5p reporter plasmid group was significantly reduced by ~70% (Fig. 3B). In another experiment, transfection of miR-1224-5p mimics reduced the levels of ZEB1-AS1 (Fig. 3C), while knockdown of ZEB1-AS1 increased the levels of miR-1224-5p (Fig. 3D). Furthermore, a negative correlation between the levels of miR-1224-5p and ZEB1-AS1 in melanoma tissues was identified (Fig. 3E). All of these results indicated that ZEB1-AS1 inhibits miR-1224-5p in melanoma.
Figure 3.
Negative association between miR-1224-5p and ZEB1-AS1. (A) A bioinformatics analysis predicted that miR-1224-5p binding sites are present in ZEB1-AS1. (B) Transfection of miR-1224-5p mimics into A375 cells inhibits the fluorescent activity of a luciferase reporter plasmid containing the WT but not the Mut miR-1224-5p-binding sequence from ZEB1-AS1. (C) Transfection of miR-1224-5p mimics reduces ZEB1-AS1 levels in A375 cells. (D) Knockdown of ZEB1-AS1 enhances miR-1224-5p levels in A375 cells. (E) A negative correlation between the levels of miR-1224-5p and ZEB1-AS1 in melanoma tissues was identified. **P<0.01, ***P<0.001 vs. NC. miR, microRNA; ZEB1-AS1, zinc finger E-box binding homeobox 1 antisense RNA 1; WT, wild-type; Mut, mutant; miR-NC, negative control miR mimics; siNC, scrambled control small interfering RNA; siZEB1-AS1, small interfering RNA #1 targeting ZEB1-AS1.
ZEB1-AS1 has a role in melanoma development by regulating miR-1224-5p
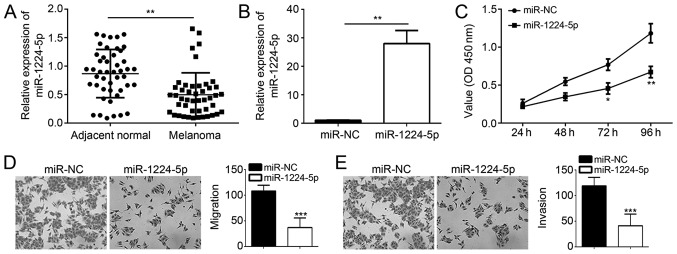
To determine whether ZEB1-AS1 promotes melanoma development via regulating miR-1224-5p, the effects of miR-1224-5p mimics on melanoma cell proliferation, invasion and migration were assessed using CCK8 and Transwell assays. The results confirmed that miR-1224-5p levels were low in melanoma vs. normal adjacent tissues (Fig. 4A). The successful transfection of miR-1224-5p mimics was confirmed by RT-qPCR (Fig. 4B). The results indicated that miR-1224-5p mimics inhibited processes of melanoma development and progression by suppressing cell proliferation, migration and invasion (P<0.05; Fig. 4C-E).
Figure 4.
miR-1224-5p mimics inhibit melanoma cell proliferation, migration and invasion. (A) miR-1224-5p expression is downregulated in melanoma tissue. (B) Confirmation of miR-1224-5p mimics transfection by RT-qPCR. (C) Transfection of miR-1224-5p mimics inhibits cell proliferation in A375 cells, as determined via the CCK8 assay. (D and E) miR-1224-5p mimics transfection significantly inhibits cell migration and invasion (magnification, ×100). *P<0.05, **P<0.01, ***P<0.001 vs. normal/NC. miR, microRNA; miR-NC, negative control miR mimics; OD, optical density.
Discussion
In the present study, the role of lncRNA ZEB1-AS1 in the pathogenesis of melanoma and the underlying molecular mechanisms were investigated. The results revealed that a negative correlation between lncRNA ZEB1-AS1 levels and the overall survival rate of melanoma patients. Furthermore, miR-1224-5p mimics reduced the levels of ZEB1-AS1, while knockdown of ZEB1-AS1 significantly increased the levels of miR-1224-5p, and a direct regulatory interaction was identified between ZEB1-AS1 and miR-1224-5p. Of note, transfection with miR-1224-5p mimics or with siZEB1-AS1 inhibited the proliferation, invasion and migration of melanoma cells.
ZEB1-AS1 may serve as a biomarker or target for cancer diagnosis or treatment, respectively, and dysregulation of ZEB1-AS1 may be a critical step in tumorigenesis and cancer progression with a potential clinical application (20). For instance, ZEB1-AS1 has an important role in the pathogenesis of bladder cancer by promoting proliferation and inhibiting apoptosis (21). Of note, downregulation of ZEB1-AS1 caused an increase in B-cell lymphoma 2 (Bcl-2)-associated X protein and a decrease in Bcl-2 expression to induce apoptosis (11). ZEB1-AS1 was also confirmed to be closely associated with metastasis and with a poor prognosis for gastric cancer patients (22). Consistent with these results, the present study indicated that the survival rate of melanoma patients with lower ZEB1-AS1 was higher. Collectively, it was suggested that ZEB1-AS1 has an important role in the genesis and progression of melanoma.
In addition to the direct regulation of miR-1224-5p, ZEB1-AS1 has been previously reported to regulate a number of further miRs in various diseases. In the pathogenesis of osteosarcoma, ZEB1-AS1 and miR-200s were indicated to have a negatively regulatory association, and dysregulation of ZEB1-AS1 to affect miR-200s contributed to the development of this disease (23). Furthermore, Zhang et al (24) reported that ZEB1-AS1 inhibits the levels of miR-335-5P to promote gastric cancer cell invasion and proliferation in vitro. Wei et al (25) reported that ZEB1-AS1 increased glioma cell migration, proliferation and invasion by regulating the expression of miR-577. All of these studies offered potential targets for disease treatment. Similarly, in the present study, ZEB1-AS1 negatively regulated the expression of miR-1224-5p to then affect the proliferation, invasion and migration of melanoma. In malignant gliomas, miR-1224-5p was previously reported to inhibit the expression of cyclic AMP responsive element binding protein 1 and reduce its tumor-promoting activity, including invasion, proliferation and apoptosis (26). Qian et al (27) also indicated that miR-1224-5p inhibits the proliferation of glioma cells, and the expression levels were associated with its clinical prognosis and grading of this disease. Of note, overexpression of miR-1224-5pM was confirmed to lead to suppression of keloid fibroblast proliferation, as well as a decrease of invasion and migration, and promotion of apoptosis in keloid fibroblasts via the transforming growth factor-β1/Smad3 signaling pathway (28). In the present study, based on bioinformatics predictions and in vitro experiments, we a negative regulatory interaction between ZEB1-AS1 and miR-1224-5p was discovered. Furthermore, knockdown of ZEB1-AS1 and transfection with miR-1224-5p mimics inhibited the proliferation, invasion and migration of melanoma. However, to better validate that ZEB1-AS1 regulates melanoma progression by inhibiting miR-1225-4p, a rescue assay by simultaneous transfection with siRNA targeting ZEB1-AS1 and miR-1224-55 mimics should be performed in the future.
In conclusion, the present study indicated that ZEB1-AS1 enhanced the proliferation, invasion and migration of melanoma cells by directly inhibiting miR-1224-5p, and overexpression of ZEB1-AS1 was also associated with a decrease in the overall survival rate of melanoma patients. These results indicated that ZEB1-AS1 and miR-1224-5p have a critical role in the pathogenesis of melanoma and may serve as a predictive biomarker and a potential target for melanoma treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
QW and DL initiated and designed the present study, analyzed the data, interpreted the results and wrote the manuscript. RZ performed several experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Regarding the use of human samples, the protocol of the present study was approved by the Institutional Ethics Committee of Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science (Xiangyang, China) and all enrolled patients signed a written informed consent document.
Patient consent for publication
All patients within the present study provide consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Levin DB, Wilson K, Valadares de Amorim G, Webber J, Kenny P, Kusser W. Detection of p53 mutations in benign and dysplastic nevi. Cancer Res. 1995;55:4278–4282. [PubMed] [Google Scholar]
- 2.Wallack MK, Sivanandham M, Balch CM, Urist MM, Bland KI, Murray D, Robinson WA, Flaherty LE, Richards JM, Bartolucci AA, et al. A phase III randomized, doúble-blind, multiinstitutional trial of vaccinia melanoma oncolysate-active specific immunotherapy for patients with stage II melanoma. Cancer. 2015;75:34–42. doi: 10.1002/1097-0142(19950101)75:1<34::AID-CNCR2820750108>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Busam KJ, Berwick M, Blessing K, Fandrey K, Kang S, Karaoli T, Fine J, Cochran AJ, White WL, Rivers J, et al. Tumor vascularity is not a prognostic factor for malignant melanoma of the skin. Am J Pathol. 1995;147:1049–1056. [PMC free article] [PubMed] [Google Scholar]
- 4.Surhone LM, Tennoe MT, Henssonow SF. Treatment and prognosis of melanoma. Betascript Publishing. 2010 [Google Scholar]
- 5.Balch CM, Murad TM, Soong SJ, Ingalls AL, Halpern NB, Maddox WA. A multifactorial analysis of melanoma: Prognostic histopathological features comparing Clark's and Breslow's staging methods. Ann Surg. 1978;188:732–742. doi: 10.1097/00000658-197812000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: A potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, Yoon CH, Novina CD. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like Region. Cell Rep. 2016;15:2025–2037. doi: 10.1016/j.celrep.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang Y, Zang W, Zhao G. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016;33:88. doi: 10.1007/s12032-016-0804-2. [DOI] [PubMed] [Google Scholar]
- 9.Yong S, Cheng H, Wang G, Yu G, Zhang D, Wang Y, Fan W, Yang W. Deregulation of miR-183 promotes melanoma development via lncRNA MALAT1 regulation and ITGB1 signal activation. Oncotarget. 2017;8:3509–3518. doi: 10.18632/oncotarget.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res. 2016;8:4095–4105. [PMC free article] [PubMed] [Google Scholar]
- 11.Lv QL, Hu L, Chen SH, Sun B, Fu ML, Qin CZ, Qu Q, Wang GH, He CJ, Zhou HH. A long noncoding RNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in Glioma. Int J Mol Sci. 2016;17(pii):E1431. doi: 10.3390/ijms17091431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su W, Xu M, Chen X, Chen N, Gong J, Nie L, Li L, Li X, Zhang M, Zhou Q. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16:142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Wang QF, Zou ML, He XA, Lv JJ. Overexpressed lncRNA ZEB1-AS1 promotes cell invasion and angiogenesis through Wnt/β-catenin signaling in non-small cell lung cancer. Int J Clin Exp Patho. 2017;10:3990–3997. [Google Scholar]
- 14.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 15.Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu P, Du Y, Wu J, Qin X, Chen R, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 16.Dalby B, Cates S, Harris A, Ohki EC, Tilkins ML, Price PJ, Ciccarone VC. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2004;33:95–103. doi: 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Dong Y, Wang X, Ma H, Sheng Z, Li G, Lu G, Sugimura H, Zhou X. Expression of EphA1 in gastric carcinomas is associated with metastasis and survival. Oncol Rep. 2010;24:1577–1584. doi: 10.3892/or_00001020. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Xiao B, Zhu ED, Li N, Lu DS, Li W, Li BS, Zhao YL, Mao XH, Guo G, Yu PW, Zou QM. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncol Rep. 2012;27:559–566. doi: 10.3892/or.2011.1514. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Li Z, Leng K, Xu Y, Ji D, Huang L, Cui Y, Jiang X. ZEB1-AS1: A crucial cancer-related long non-coding RNA. Cell Prolif. 2018;51 doi: 10.1111/cpr.12423. doi: 10.1111/cpr.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Zhan Y, Liu Y, Chen Z, Liang J, Li W, He A, Zhou L, Mei H, Wang F, Huang W. Increased expression of ZEB1-AS1 correlates with higher histopathological grade and promotes tumorigenesis in bladder cancer. Oncotarget. 2017;8:24202–24212. doi: 10.18632/oncotarget.15527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu XJ, Li SL, Li JS, Lu H, Yin LL, Zheng WF, Wang WC. Long non-coding RNA ZEB1-AS1 is associated with poor prognosis in gastric cancer and promotes cancer cell metastasis. Eur Rev Med Pharmacol Sci. 2018;22:2624–2630. doi: 10.26355/eurrev_201805_14956. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Pan C, Cai Y, Wang H. Interplay between long noncoding RNA ZEB1-AS1 and miR-200s regulates osteosarcoma cell proliferation and migration. J Cell Biochem. 2017;118:2250–2260. doi: 10.1002/jcb.25879. [DOI] [PubMed] [Google Scholar]
- 24.Zhang LL, Zhang LF, Guo XH, Zhang DZ, Yang F, Fan YY. Downregulation of miR-335-5p by long noncoding RNA ZEB1-AS1 in gastric cancer promotes tumor proliferation and invasion. DNA Cell Biol. 2018;37:46–52. doi: 10.1089/dna.2017.3926. [DOI] [PubMed] [Google Scholar]
- 25.Wei N, Wei H, Zhang H. Long non-coding RNA ZEB1-AS1 promotes glioma cell proliferation, migration and invasion through regulating miR-577. Eur Rev Med Pharmacol Sci. 2018;22:3085–3093. doi: 10.26355/eurrev_201805_15068. [DOI] [PubMed] [Google Scholar]
- 26.Jin Q, Rui L, Wang YY, Shi Y, Luan WK, Tao T, Zhang JX, Xu YC, You YP. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem. 2015;403:33–41. doi: 10.1007/s11010-015-2334-1. [DOI] [PubMed] [Google Scholar]
- 27.Qian J, Li R, Wang YY, Luan WK, Tao T, Zhang JX, Xu YC, You YP. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem. 2015;403:33–41. doi: 10.1007/s11010-015-2334-1. [DOI] [PubMed] [Google Scholar]
- 28.Yao X, Cui X, Wu X, Xu P, Zhu W, Chen X, Zhao T. Tumor suppressive role of miR-1224-5p in keloid proliferation, apoptosis and invasion via the TGF-β1/Smad3 signaling pathway. Biochem Biophys Res Commun. 2018;495:713–720. doi: 10.1016/j.bbrc.2017.10.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.