Abstract
Long non-coding RNAs (lncRNAs) serve an essential role in regulating immunological functions. However, their impact on Henoch-Schönlein purpura nephritis (HSPN), has remained elusive. The present study determined the expression of lncRNAs and mRNAs in the peripheral blood of 6 children with HSPN and recruited 4 healthy children for comparative study. High-throughput sequencing revealed outstanding differences in lncRNA and mRNA expression, which were verified through reverse transcription-quantitative polymerase chain reaction. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses were used to investigate the associated biological functions and possible mechanisms of lncRNAs and mRNAs in HSPN. A total of 820 differentially expressed lncRNAs between the two groups were identified, of which 34 were upregulated and 786 were downregulated. Simultaneously, a total of 3,557 mRNAs were also identified to be differentially expressed, of which 1,232 were upregulated and 2,325 were downregulated. The results revealed that the expression of lncRNAs including ENST00000378432, ENST00000571370, uc001kfc.1 and uc010qna.2 was decreased in HSPN patients compared with that in healthy controls. These lncRNAs were associated with the p53 signaling pathway and apoptosis-associated genes (AKT2, tumor protein 53, phosphatase and tensin homolog and FAS). Further studies of those lncRNAs will be performed to elucidate their functions in apoptosis. Complete raw data files were deposited in the Gene Expression Omnibus (GEO) at National Center for Biotechnology information under the GEO accession no. GSE102114 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102114).
Keywords: Henoch-Schönlein purpura, Henoch-Schönlein purpura nephritis, long non-coding RNA, mRNA, high-throughput sequencing analysis
Introduction
Henoch-Schönlein purpura (HSP) is the most common type of systemic small-vessel vasculitis in pediatric patients, and is associated with multiple and complex pathogenic factors, as well as diverse pathological damage (1). It is a systemic disorder characterized by leukocytoclastic vasculitis involving the capillaries and deposition of immunoglobulin (Ig)A immune complexes (2). Of all pediatric patients with HSP, >90% are <10 years old (3,4). Between weeks 4 and 6 of the initial disease presentation, ~40% of pediatric patients with HSP progress to HSP nephritis (HSPN), which is one of the major manifestations and the primary cause of mortality associated with HSP (2,5,6). To date, however, the exact pathophysiology of HSPN has remained largely elusive and requires further investigation.
Long non-coding RNAs (lncRNAs) are a class of RNA with a length of >200 nucleotides and no coding function. In recent years, as the rapid development of technologies has facilitated the analysis of the ‘transcriptome’ the study of lncRNAs has enabled the discovery of comprehensive genetic information in the human genome. A large amount of evidence has suggested that lncRNAs regulate protein-coding genes at the transcriptional and post-transcriptional levels, and exert transcription control (7,8). Numerous studies have provided novel insight into different expression profiles of lncRNAs in a number of human kidney diseases, including acute kidney rejection, diabetic nephropathy, membranous nephropathy, chronic kidney disease and lupus nephritis (9–15). In parallel with their role in disease pathogenesis, lncRNAs may also serve as a potential biomarker of disease status and aid in the diagnosis, prognosis and clinical management of disease (16,17). However, the expression patterns and functions of lncRNAs in HSPN have largely remained to be elucidated.
In the present study, high-throughput sequencing was applied to identify 820 lncRNAs and 3,557 mRNAs that are significantly aberrantly expressed in the peripheral blood of pediatric patients with HSPN. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) provided results that were consistent with those obtained by data analysis of the gene expression profiles, thereby verifying them. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were then performed to elucidate the roles and pathways of the differentially expressed RNAs. These results indicated that the aberrantly expressed RNAs may have important roles in the development of HSPN through promoting serum proteins generation and regulating the apoptosis pathway, and that knowing the differently expressed RNAs might provide useful biomarkers for HSPN therapy and diagnosis.
Patients and methods
Patients and sample collection
A total of 6 pediatric patients with HSPN (4 males and 2 females; mean age, 12.17±1.72 years) were recruited at the Affiliated Hospital of Liaoning University of Traditional Chinese Medicine (Shenyang, China) between November 2016 and March 2017. The diagnostic criteria for HSPN were according to those outlined at the Congress of the Chinese Pediatric Society in 2000 (18). Patients with other coexisting renal pathologies were excluded. None of the patients of the present study was diagnosed with any other complications. Furthermore, none of the subjects had taken any hormonal or immunosuppressive drugs for ≥6 months prior to the commencement of the study. The clinical characteristics of the patients with HSPN are presented in Table I. The mean urine red blood cell count, 24-h urine protein quantity and serum IgA were 387.48±590.34 p/µl, 0.28±0.15 g/24 h and 3.00±1.21 g/l, respectively. A total of 4 age matched healthy subjects (1 male and 3 females; mean age, 11.25±2.99 years) were selected as healthy controls (HC). Blood samples were obtained from the 6 children with HSPN and 4 healthy volunteers.
Table I.
Clinical characteristics of 6 HSPN cases.
| Patient ID | S64 | S87 | S105 | S111 | S173 | S303 |
|---|---|---|---|---|---|---|
| Age (years) | 14 | 13 | 10 | 14 | 11 | 11 |
| Sex | F | F | M | M | M | M |
| Purpura | + | + | + | + | + | + |
| Abdominal pain | − | + | − | − | − | + |
| Arthralgia | − | + | + | − | + | + |
| Period between the onset of HSPN | 12 | 10 | 34 | 16 | 60 | 20 |
| and initiation of therapy (months) |
HSPN, Henoch-Schönlein purpura nephritis; S, sick; F, female; M, male.
The present study was approved by the Ethics Committee of the Institutional Ethics Board of the Affiliated Hospital of Liaoning University of Traditional Chinese Medicine (approval no. 2016CS(KT)-002-01). Written informed consent was obtained from the parents of all of the pediatric subjects enrolled in the present study.
RNA extraction and library construction
Peripheral blood was obtained from the subjects of the groups and collected in tubes containing EDTA as an anticoagulant. Peripheral blood total RNA was isolated using a total RNA isolation kit purchased from BioTeke Corp. (Beijing, China), according to the manufacturer's protocols. From the RNA (1 µg) the ribosomal (r)RNA was removed using a Ribo-Zero rRNA Removal kit (Illumina, Inc., San Diego, CA, USA), according to the manufacturer's protocols. RNA libraries were constructed using rRNA-depleted RNAs with a TruSeq Stranded Total RNA Library Prep kit (Illumina, Inc.), according to the manufacturer's protocols. Libraries were controlled for quality and were quantified using a BioAnalyzer 2100 system (Agilent Technologies, Inc., Santa Clara, CA, USA).
High-throughput sequencing and computational analysis
A DNA library (420 µl of a 10 pM library) was denatured as single-stranded DNA molecules, captured on Illumina flow cells, amplified in situ as clusters and finally sequenced for 150 cycles on an Illumina Hiseq Sequencer, according to the manufacturer's protocols. High-throughput sequencing was performed by Shanghai Cloud-Seq Biotech, Inc., (Shanghai, China). In brief, paired-end reads were harvested from the Illumina HiSeq 2000 sequencer (Illumina, Inc.), and were quality controlled via their Q score (>Q30). Following 3′ adaptor-trimming and removal of low-quality reads with cutadapt software (v1.9.3) (19), the high-quality, trimmed reads were aligned to a reference genome (UCSC HG19) guided by the Ensembl GFF gene annotation file with hisat2 software (v2.0.4; http://ccb.jhu.edu/software/hisat2/index.shtml). Next, cuffdiff software (v2.2.1, part of cufflinks; http://cufflinks.cbcb.umd.edu/) was used to obtain the gene fragment counts per kilobase of exon per million fragments mapped (FPKM) as the expression profiles of lncRNA and mRNA, and the fold change (FC) and P-value were calculated based on the FPKM, from which differentially expressed lncRNAs and mRNAs were identified.
GO term analysis and pathway analysis
The GO (www.geneontology.org) and KEGG (www.genome.ad.jp/kegg) databases were utilized to analyze biological functions and signaling pathways based on the differentially expressed lncRNAs and mRNAs. Fisher's exact test was used to determine whether there was more overlap between the gene list and the GO annotation list than that expected to occur by chance. The P-value denoted the significance of the GO term enrichment and was the basis of the significance level, with P<0.05 set as the threshold. Pathway analysis was used to investigate the major pathways of the aberrantly expressed genes according to the KEGG database. The threshold for the false discovery rate was set at 0.05, and P<0.05 was considered to indicate a statistically significant difference.
RT-qPCR validation
Total RNA was extracted from the peripheral blood using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocols. In brief, 1 µg total RNA from each sample was used for the synthesis of first strand complementary DNA using a SuperScript™ III First-Strand Synthesis System kit (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. qPCR was performed on an Applied Biosystems ViiA™ 7 Real-time PCR System using the SYBR-Green method (both Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The thermocycling conditions were as follows: A denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. The primer sequences are presented in Table II. Relative gene expression levels were quantified using the 2−∆∆Cq method, in which β-actin (ACTB) was used as an internal control (20).
Table II.
Primers used for reverse transcription-quantitative polymerase chain reaction.
| Gene name/direction | Sequence (5′ to 3′) | Product length (bp) |
|---|---|---|
| ENSG00000267121 | ||
| F | GAGGAAGACCCTGGAAGGAG | 203 |
| R | GTCCCAAGCTTCAGTCATCC | |
| ENSG00000252310 | ||
| F | GGTCCGAGTGTTGTGGGTTA | 50 |
| R | GGGGGAGACAATGTTAAATCAA | |
| uc001kfc.1 | ||
| F | AAAATTAGCCAGGCATGGTG | 209 |
| R | TCTCTCACGGCTCTTGTGTG | |
| uc010qna.2 | ||
| F | GGGTCTTCCTCATGGCACTA | 202 |
| R | CAGGCCTTCCAAGTTCTGAG | |
| ENST00000378432 | ||
| F | CCTTTTCTCCATGGCATTTG | 202 |
| R | TCCTGCATTCATTCATTCCA | |
| ENST00000571370 | ||
| F | GGTTGTTTCATTCCGCAGTT | 204 |
| R | TTTCTGGGACGATGAAAAGG | |
| ACTB | ||
| F | GGCCTCCAAGGAGTAAGACC | 73 |
| R | AGGGGAGATTCAGTGTGGTG |
F, forward; R, reverse; ACTB, β actin.
Statistical analysis
Statistical analysis and graphic presentation were performed with SPSS version 19.0 software packages (IBM Corp., Armonk, NY, USA). Measurement data are expressed as the mean ± standard deviation. The differences in expression levels of tested lncRNAs and mRNAs between two groups were assessed using Student's t-test. P<0.05 was considered to indicate a statistical significant difference. FC≥1.5 indicated upregulation and <0.67 indicated downregulation. were considered to indicate significant differences. Otherwise, the non-parametric Mann-Whitney U test was used to analyze the data. Fisher's exact test was used for GO analysis and KEGG pathway analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Subject characteristics
The baseline demographic and clinical data of the subjects in the HSPN group and the HC group are summarized in Table III. The pediatric patients with HSPN had a significantly higher urine red blood cell count, 24-h urine protein quantity and serum IgA than the HC group.
Table III.
Baseline demographic and clinical data of subjects.
| Parameter | HSPN group (n=6) | HC group (n=4) | Normal range |
|---|---|---|---|
| Sex (male/female), n | 4:2 | 1:3 | |
| Age (years, mean ± SD) | 12.17±1.72 | 11.25±2.99 | |
| Serum IgA (g/l, mean ± SD) | 3.00±1.21 | 1.39±0.55 | 0.63–1.79 |
| Urine red blood cell count (p/µl, mean ± SD) | 387.48±590.34 | 3.60±8.10 | 0-25.00 |
| 24-h urine protein quantity (g/24 h, mean ± SD) | 0.280±0.150 | 0.050±0.007 | 0–0.150 |
HSPN, Henoch-Schönlein purpura nephritis; HC, healthy controls; SD, standard deviation.
Differentially expressed lncRNAs
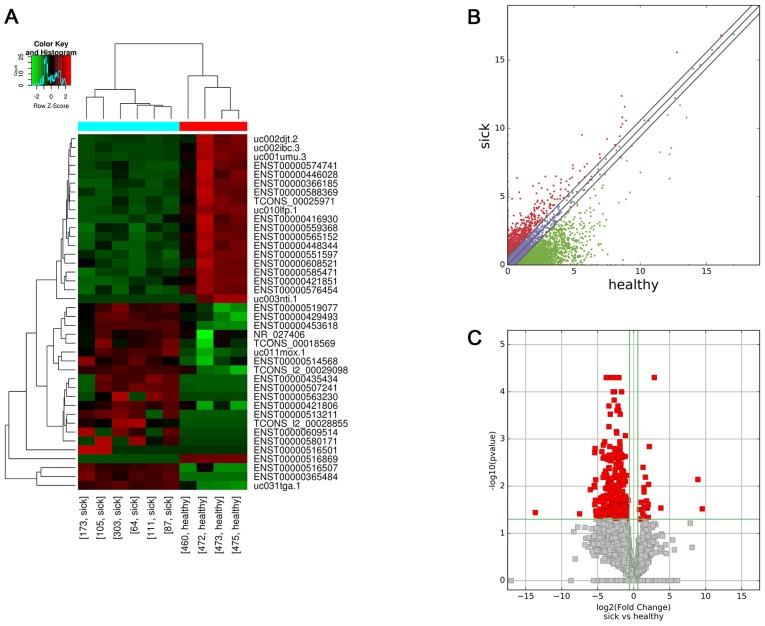
A total of 820 lncRNAs were aberrantly expressed between the HSPN group and the HC group. High-throughput sequencing revealed that 34 lncRNAs were upregulated and 786 lncRNAs were downregulated. The top 25 differentially expressed lncRNAs according to their FC values in the HSPN vs. HC group are presented in Table IV. Hierarchical clustering was performed according to the lncRNA expression levels in these 10 samples (Fig. 1A). Scatter and volcano plots were used to assess variations in lncRNA expression between the two groups (Fig. 1B and C, respectively).
Table IV.
Top 25 aberrantly expressed lncRNAs according to the FC values between the two groups.
| lncRNA ID | FC value | P-value | Class | Database |
|---|---|---|---|---|
| Upregulated | ||||
| ENSG00000202354 | 736.95 | 0.030 | Intergenic | Ensembl |
| ENSG00000252310 | 487.59 | 0.007 | Intergenic | Ensembl |
| Downregulated | ||||
| ENSG00000259001 | −12971.66 | 0.036 | Bidirectional | Ensembl |
| HLA-B | −182.99 | 0.039 | Sense | UCSC_knowngene |
| ENSG00000257621 | −65.40 | 0.012 | Intronic | Ensembl |
| ENSG00000262380 | −44.54 | 0.010 | Sense | Ensembl |
| BC047651 | −42.38 | 0.010 | Sense | UCSC_knowngene |
| ENSG00000267121 | −42.31 | 0.002 | Intergenic | Ensembl |
| ENSG00000230105 | −42.22 | 0.025 | Intergenic | Ensembl |
| DQ598910 | −41.06 | 0.002 | Sense | UCSC_knowngene |
| ENSG00000236535 | −39.99 | 0.020 | Sense | Ensembl |
| ENSG00000262879 | −38.70 | 0.007 | Intergenic | Ensembl |
| BC044596 | −38.50 | 0.034 | Sense | UCSC_knowngene |
| ENSG00000231485 | −36.49 | 0.041 | Intergenic | Ensembl |
| ENSG00000260060 | −26.54 | 0.045 | Intergenic | Ensembl |
| ENSG00000232593 | −26.14 | 0.031 | Intergenic | Ensembl |
| ENSG00000246016 | −24.65 | 0.018 | Intergenic | Ensembl |
| MGC72080 | −24.52 | 0.021 | Intergenic | UCSC_knowngene |
| XLOC_012288 | −23.80 | 0.036 | Intergenic | TCONS |
| ENSG00000247556 | −21.99 | 0.018 | Intergenic | Ensembl |
| ENSG00000267672 | −21.24 | 0.002 | Sense | Ensembl |
| ENSG00000227195 | −19.36 | 0.035 | Intergenic | Ensembl |
| AX747098 | −19.05 | 0.018 | Sense | UCSC_knowngene |
| AK098491 | −18.85 | 0.007 | Sense | UCSC_knowngene |
| ENSG00000253430 | −18.81 | 0.003 | Sense | Ensembl |
FC, fold-change; lncRNA, long non-coding RNA; HLA-B, human leukocyte antigen-B.
Figure 1.
Profiles of differentially expressed lncRNAs between the two groups. (A) Hierarchical clustering analysis of 40 lncRNAs that were aberrantly expressed between children with HSPN (sick173, sick105, sick303, sick64, sick111 and sick87) and HC (healthy460, healthy472, healthy473 and healthy475). Red and green indicate high and low relative expression, respectively. P<0.05 was considered to indicate a statistical significant difference. FC≥1.5 indicated upregulation and <0.67 indicated downregulation. (B) Scatter plot of aberrant expression of lncRNAs between the HSPN and HC libraries. Red indicates upregulated lncRNAs [log2(HSPN/HC)>0], blue indicates lncRNAs with no change in expression between HSPN and HC libraries, green indicates downregulated lncRNAs [log2(HSPN/HC)<0]. (C) Volcano plot of aberrant lncRNA expression. The vertical green lines delineate a 1.5-fold upregulation and 0.67-fold downregulation of lncRNAs, and the horizontal line represents P=0.05. Red plots on the right side represent 34 upregulated lncRNAs with FC≥1.5 and corrected P<0.05. Red plots on the left side represent 786 down-regulated lncRNAs with FC<0.67 and corrected P<0.05. lncRNA, long non-coding RNA; HSPN, Henoch-Schönlein purpura nephritis; HC, healthy controls; FC, fold change; NR, nitrate reductase.
Differentially expressed mRNAs
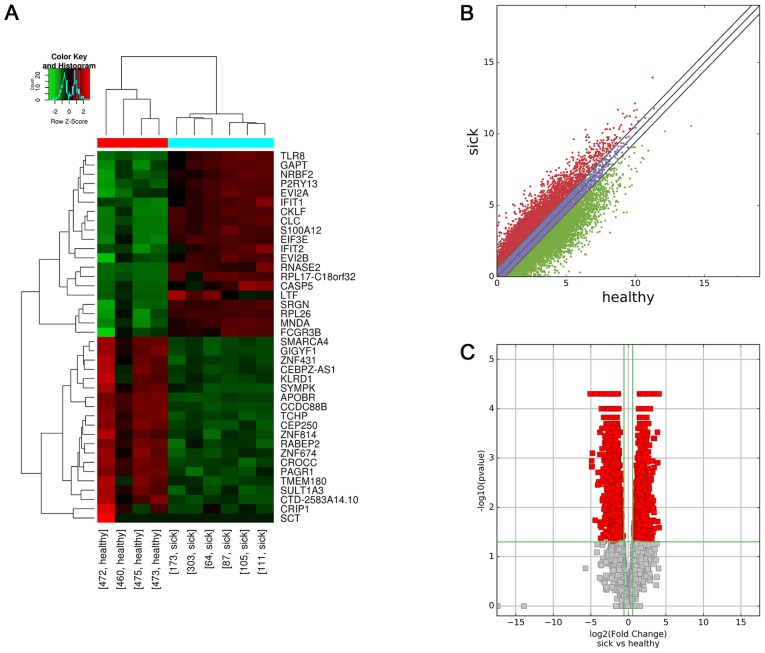
A total of 3,557 mRNAs that were aberrantly expressed between the two groups were identified using the same criteria as those for the lncRNAs. A total of 1,232 upregulated and 2,325 downregulated differentially expressed mRNAs were identified. The top 25 differentially expressed mRNAs are listed in Table V. Hierarchical clustering was performed according to the mRNA expression levels in all 10 samples (Fig. 2A). Scatter and volcano plots generated from these differentially expressed mRNAs exhibited a clear segregation between the HSPN and HC groups (Fig. 2B and C, respectively).
Table V.
Top 25 differentially expressed mRNAs according to the FC values between the two groups.
| Gene name | Ensembl ID | FC value | P-value | Chromosome |
|---|---|---|---|---|
| Upregulated | ||||
| RPL17-C18orf32 | ENSG00000215472 | 17.16 | 0.026 | chr18:47008027-47018906 |
| MNDA | ENSG00000163563 | 16.94 | <0.001 | chr1:158801106-158819296 |
| IFIT1 | ENSG00000185745 | 15.09 | 0.001 | chr10:90973325-91174314 |
| Downregulated | ||||
| ZNF431 | ENSG00000196705 | −33.88 | <0.001 | chr19:21324826-21373034 |
| APOBR | ENSG00000184730 | −29.51 | 0.001 | chr16:28467692-28510291 |
| CROCC | ENSG00000058453 | −29.46 | 0.002 | chr1:17066767-17299474 |
| CTD-2583A14.10 | ENSG00000268750 | −28.25 | 0.002 | chr19:58281022-58427978 |
| PAGR1 | ENSG00000263136 | −27.74 | 0.001 | chr16:29262828-30215631 |
| KLRD1 | ENSG00000134539 | −25.48 | <0.001 | chr12:10378656-10469850 |
| SYMPK | ENSG00000125755 | −21.33 | <0.001 | chr19:46318667-46366548 |
| SCT | ENSG00000070031 | −21.32 | 0.018 | chr11:626430-627143 |
| CEP250 | ENSG00000126001 | −20.73 | <0.001 | chr20:34042984-34099804 |
| ZNF674 | ENSG00000251192 | −20.15 | <0.001 | chrX:46357161-46404892 |
| GIGYF1 | ENSG00000146830 | −19.36 | <0.001 | chr7:100277129-100287071 |
| ZNF814 | ENSG00000204514 | −18.82 | <0.001 | chr19:58281022-58427978 |
| CEBPZ-AS1 | ENSG00000218739 | −18.02 | <0.001 | chr2:37394962-37551951 |
| RABEP2 | ENSG00000177548 | −17.18 | <0.001 | chr16:28889725-28950667 |
| SMARCA4 | ENSG00000127616 | −17.17 | <0.001 | chr19:11071597-11176071 |
| CRIP1 | ENSG00000257341 | −16.97 | 0.004 | chr14:105952653-105965912 |
| SULT1A3 | ENSG00000261052 | −16.30 | 0.002 | chr16:29262828-30215631 |
| TMEM180 | ENSG00000138111 | −15.97 | <0.001 | chr10:104221148-104236802 |
| CCDC88B | ENSG00000168071 | −15.71 | <0.001 | chr11:64107694-64125006 |
| TCHP | ENSG00000139437 | −15.38 | <0.001 | chr12:110338068-110434194 |
| CHCHD6 | ENSG00000159685 | −15.37 | 0.003 | chr3:126423062-126679249 |
| BAIAP2 | ENSG00000175866 | −15.31 | <0.001 | chr17:79008947-79156964 |
FC, fold-change; MNDA, myeloid nuclear differentiation antigen; IFIT1, interferon-induced protein with tetratricopeptide repeats 1; ZNF431, zinc finger protein 431; APOBR, apolipoprotein B receptor; CROCC, ciliary rootlet coiled-coil; PAGR1, PAXIP1 associated glutamate rich protein 1; KLRD1, killer cell lectin like receptor D1; SYMPK, symplekin; SCT, secretin; CEP250, centrosomal protein 250; ZNF674, zinc finger protein 674; GIGYF1, GRB10 interacting GYF protein 1; ZNF814, zinc finger protein 814; CEBPZ-AS1, CEBPZ opposite strand; RABEP2, RAB GTPase binding effector protein 2; SMARCA4, SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4; CRIP1, cysteine rich protein 1; SULT1A3, sulfotransferase family 1A member 3; TMEM180, major facilitator superfamily domain containing 13A; CCDC88B, coiled-coil domain containing 88B; TCHP, trichoplein keratin filament binding; CHCHD6, coiled-coil-helix-coiled-coil-helix domain containing 6; BAIAP2, BAI1 associated protein 2; chr, chromosome.
Figure 2.
Differential expression of mRNAs between the two groups. (A) Hierarchical clustering analysis of 40 differentially expressed mRNAs from each sample. FC≥1.5 indicated upregulation and <0.67 indicated downregulation. (B) Scatter plot of differential expression of mRNAs between the HSPN and HC libraries. Red indicates upregulated mRNAs [log2(HSPN/HC)>0], blue indicates mRNAs with no change in expression between HSPN and HC libraries, green indicates downregulated mRNAs [log2(HSPN/HC)<0]. (C) Volcano plot of differential mRNA expression. The vertical green lines delineate a 1.5-fold upregulation and 0.67-fold downregulation of mRNAs, and the horizontal line represents P=0.05. Red plots on the right side represent 1,232 upregulated mRNAs with FC≥1.5 and corrected P<0.05. Red plots on the left side represent 2,325 downregulated mRNAs with FC<0.67 and corrected P<0.05. HSPN, Henoch-Schönlein purpura nephritis; HC, healthy controls; FC, fold change.
Validation of sequencing data using RT-qPCR
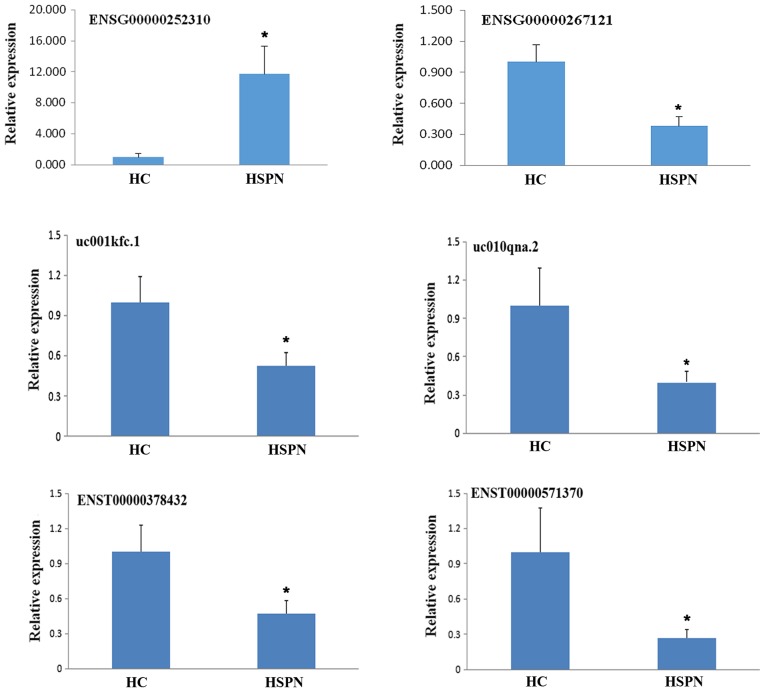
In order to validate the sequencing analysis, 6 differentially expressed lncRNAs were selected. RT-qPCR analysis verified that the lncRNA ENSG00000252310 was upregulated, while ENSG00000267121, uc001kfc.1, uc010qna.2, ENST00000378432 and ENST00000571370 were downregulated in the 10 samples (Fig. 3). It was identified that the RT-qPCR results were consistent with the high-throughput sequencing data.
Figure 3.
Validation of 6 randomly selected differentially expressed lncRNAs in 10 samples by reverse transcription-quantitative polymerase chain reaction. Compared with the HC, ENSG00000252310 was upregulated, and ENSG00000267121, uc001kfc.1, uc010qna.2, ENST00000378432 and ENST00000571370 were downregulated in children with HSPN. The results were consistent with those of the high-throughput sequencing analysis. *P<0.05 vs. HC. HSPN, Henoch-Schönlein purpura nephritis; HC, healthy controls.
GO and KEGG pathway enrichment analysis
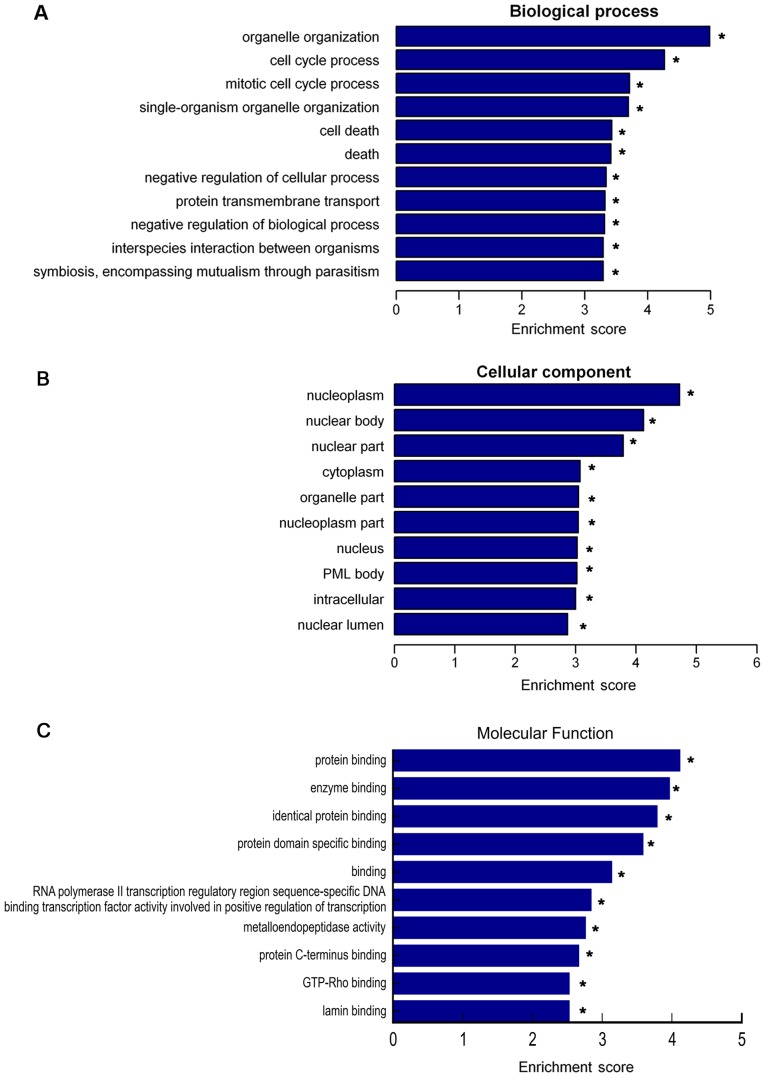
To investigate potential lncRNAs and the moieties that they regulate, which are enriched in the GO terms biological process, cellular component and molecular function, GO analysis was performed for the differentially expressed lncRNAs (Fig. 4). The present study focused on GO terms enriched among downregulated lncRNAs, and the top 10 terms in the category biological process were as follows: i) Organelle organization, ii) cell cycle process, iii) mitotic cell cycle process, iv) single-organism organelle organization, v) cell death, vi) death, vii) negative regulation of cellular process, viii) protein transmembrane transport, ix) negative regulation of biological process and x) interspecies interaction between organisms (Fig. 4A). The top 10 GO terms in the category cellular component were as follows: i) Nucleoplasm, ii) nuclear body, iii) nuclear part, iv) cytoplasm, v) organelle part, vi) nucleoplasm part, vii) nucleus, viii) promyelocytic leukemia protein body, ix) intracellular and x) nuclear lumen (Fig. 4B). The top 10 GO terms in the category molecular function were as follows: i) Protein binding, ii) enzyme binding, iii) identical protein binding, iv) protein domain specific binding, v) binding, vi) RNA polymerase II transcription regulatory region sequence-specific DNA binding transcription factor activity involved in positive regulation of transcription, vii) metalloendopeptidase activity, viii) protein C-terminus binding, ix) GTP-Rho binding and x) lamin binding (Fig. 4C).
Figure 4.
Gene Ontology analysis of distinctively expressed long non-coding RNAs in children with HSPN. (A) Top 11 terms in the category biological processes. (B) Top 10 terms in the category cellular components. (C) Top 10 terms in the category molecular function. *P<0.05 vs. HC. HSPN, Henoch-Schönlein purpura nephritis; HC, healthy controls; PML, promyelocytic leukemia protein.
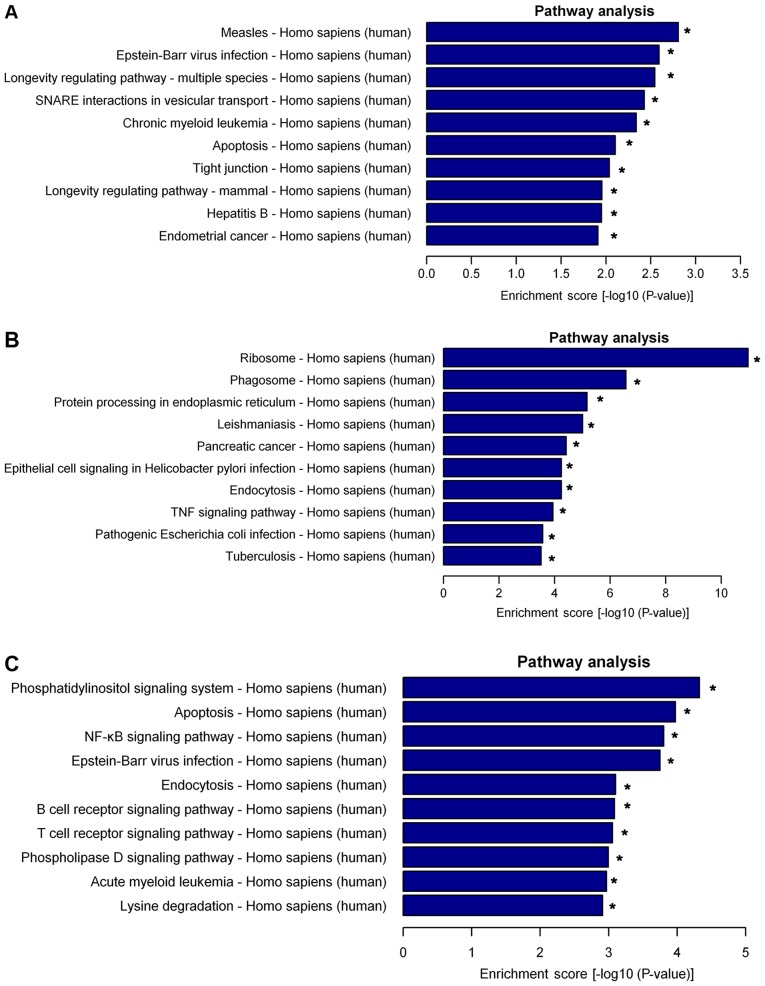
Pathway analysis indicated that 25 pathways were significantly enriched among the differentially expressed lncRNAs. The top 10 significantly enriched pathways were presented as follows: i) Measles, ii) Epstein-Barr virus infection, iii) longevity regulating pathway-multiple, iv) soluble N-ethylmaleimide sensitive factor attachment protein receptor interactions in vesicular transport, v) chronic myeloid leukemia, vi) apoptosis, vii) tight junction, viii) longevity regulating pathway-mammal, ix) hepatitis B and x) endometrial cancer (Fig. 5A). Furthermore, the differently expressed mRNAs were also subjected to KEGG analysis, the top 10 enriched pathways for the upregulated mRNAs were presented as follows: i) Ribosome, ii) phagosome, iii) protein processing in endoplasmic reticulum, iv) leishmaniasis, v) pancreatic cancer, vi) epithelial cell signaling in Helicobacter pylori infection, vii) endocytosis, viii) tumor necrosis factor (TNF) signaling pathway, ix) pathogenic Escherichia coli infection and x) tuberculosis (Fig. 5B). The top 11 enriched pathways for the downregulated mRNAs were presented as follows: i) Phosphatidylinositol signaling system, ii) apoptosis, iii) nuclear factor (NF)-κB signaling pathway, iv) Epstein-Barr virus infection, v) endocytosis, vi) B cell receptor signaling pathway, vii) T cell receptor signaling pathway, viii) T cell receptor signaling pathway, ix) phospholipase D signaling pathway, x) acute myeloid leukemia and xi) lysine degradation (Fig. 5C).
Figure 5.
Pathway analysis of differentially expressed lncRNAs and mRNAs in children with HSPN. (A) Top 10 enriched pathways among the aberrantly expressed lncRNAs in the HSPN group. (B) Top 10 enriched pathways among the upregulated mRNAs in the HSPN group. (C) Top 10 pathways among all of the downregulated mRNAs. *P<0.05 vs. HC. lncRNA, long non-coding RNA; HSPN, Henoch-Schönlein purpura nephritis; HC, healthy controls; SNARE, soluble N-ethylmaleimide sensitive factor attachment protein receptor; TNF, tumor necrosis factor; NF-κB, nuclear factor-κB.
Discussion
HSPN is a major public health problem that accounts for 78.9% of secondary glomerulopathies in pediatric patients (21). Etiologically, persistent purpura or relapse, severe abdominal symptoms (abdominal pain, gastrointestinal bleeding and severe bowel angina), arthritis and being aged >10 years old are the most significant risk factors for pediatric HSPN (22–24). Initially, lncRNAs were assumed to simply be leaky transcription noise (25). However, an abundance of studies have demonstrated that lncRNAs serve important roles in physiological processes and the pathophysiology of numerous diseases, participating in the regulation of DNA methylation, histone modification, basal transcription, post-transcriptional processes, directly binding proteins and protein function (26–30).
To the best of our knowledge, the present study was the first to investigate the expression profile of lncRNAs and mRNAs in patients with HSPN. Total RNA was extracted from the peripheral blood of patients and HC, and differentially expressed lncRNAs and mRNAs were identified using high-throughput sequencing, followed by verification of certain RNAs by RT-qPCR analysis. In the RT-qPCR experiments, ACTB was used as an endogenous control for normalising the expression of lncRNAs and mRNAs according to previous studies (31–34). Studies have demonstrated that ACTB is suitable reference gene in gene expression studies of human diseases when human peripheral blood as samples (35–37), and may be more stably expressed in whole blood than in peripheral blood mononuclear cells (38). In the present study, it was identified that the RT-qPCR results were consistent with the high-throughput sequencing data.
A total of 820 lncRNAs, including 34 upregulated and 786 downregulated ones, were identified to be differentially expressed between the two groups. In addition, 3,557 differentially expressed protein-coding mRNAs were identified from the same samples, including 1,232 upregulated and 2,325 downregulated mRNAs. The results demonstrated that the expression of lncRNAs and mRNAs in patients with HSPN were quite different from that in HC. Additionally, a greater number of lncRNAs and mRNAs were significantly downregulated than upregulated in patients with HSPN. GO and KEGG pathway analyses were used to investigate the possible mechanistic roles and pathways of lncRNAs and mRNAs in HSPN.
An integrative method involving pathway analysis was applied to identify possible functional associations between the different RNA molecules. Based on the differentially expressed mRNAs, a pathway analysis revealed via which biological functions and mechanisms they may be involved in HSPN formation. HSPN is a small-vessel form of autoimmune vasculitis caused by IgA1-mediated inflammation (39,40). The results of the present study suggested that a variety of KEGG pathways, including ribosomal function, phagosome activity, protein processing in the endoplasmic reticulum and endocytosis, are significantly enriched among the upregulated mRNAs from patients with HSPN. The majority of these pathways are involved in the generation of serum proteins, including IgA. Among these associated pathways, epithelial cell signaling in Helicobacter pylori infection, tumor necrosis factor TNF signaling, pathogenic Escherichia coli infection, the phosphatidylinositol signaling system, NF-κB signaling, Epstein-Barr virus infection, the B cell receptor signaling pathway and the T cell receptor signaling pathway exhibited significant changes in upregulated and downregulated mRNAs. In line with this, Shin et al (41) reported that Helicobacter pylori infection may cause the serum levels of IgA, C3 and cryoglobulins to increase, which promotes immune complex formation and increases the risk of HSP occurrence. Hirayama et al (42) demonstrated that the percentage values of the CD3-gated β-chain of the T cell receptor in patients with HSPN were significantly increased compared with those in healthy individuals. Chen et al (43) revealed that TNF-like weak inducer of apoptosis, a member of the TNF family, was elevated in patients with acute-stage HSP and may act as a regulator of NF-κB activation and chemokine production in human dermal microvascular endothelial cells, promoting leucocyte migration in cutaneous vasculitis. These results indicated that aberrant IgA circulating immune complexes, as well as elevated pro-inflammatory cytokines and chemokines, are all associated with the pathogenesis of HSPN.
Previous studies have demonstrated that apoptosis is one of the most important factors for controlling inflammation in inflammatory diseases such as HSP (44,45). Among the top 10 enriched pathways of the aberrantly expressed lncRNAs in the HSPN group, the present study focused on the apoptotic pathway, which serves an important role in the pathogenesis of HSPN. Yuan et al (46) suggested that IgA1 from patients with HSP may induce apoptosis of human umbilical vein endothelial cells, which may be associated with vascular endothelial injury in HSP. Ozaltin et al (45) observed a marked expression of Fas on peripheral blood neutrophils and lymphocytes in patients with HSP in the acute and the resolution phases. This suggested that increased removal of inflammatory cells through apoptosis may contribute to the early control of the inflammatory response and repair in this self-limited vasculitis. The present study revealed that ENST00000378432, ENST00000571370, uc001kfc.1 and uc010qna.2 expression was decreased in patients with HSPN and healthy controls. These lncRNAs were associated with the p53 signaling pathway and apoptosis-associated genes (AKT serine/threonine kinase 2, tumor protein 53, phosphatase and tensin homolog and FAS). Further studies on these differentially expressed lncRNAs will be performed to establish their functions in apoptosis.
The major limitation of the present study was the sample size of the patients and controls. The present results should be validated in larger cohorts. Interaction networks analyses are also required to further investigate the associations between ncRNAs, coding RNAs and proteins. Furthermore, lncRNAs and mRNAs validated to be associated with HSPN by RT-qPCR should be further investigated at the cellular level.
In conclusion, the results of the present study demonstrated a significant difference in the expression of certain lncRNAs and mRNAs between patients with HSPN and HCs. The results indicated that lncRNAs are important regulators in HSPN pathophysiological mechanisms. Functional research on these lncRNAs may be a novel and interesting research field.
Acknowledgements
The authors of the present study would like to thank Shanghai Cloud-Seq Biotech Laboratory, for their help in the guidance of the experiments and analysis of the data of this manuscript.
Glossary
Abbreviations
- HSPN
Henoch-Schönlein purpura nephritis
- lncRNA
long non-coding RNA
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- FPKM
fragment per kilobase of exon per million fragments mapped
- HC
healthy controls
- ACTB
β-actin
Funding
The present study was financially supported by the 2015 National Scientific Research Specific of Traditional Chinese Medicine Industry (grant no. 201507001-03). The State Administration of Traditional Chinese Medicine of the People's Republic of China contributed to the conception of the study and helped perform the collection of data.
Availability of data and materials
The datasets generated and analyzed during the present study are available from the Gene Expression Omnibus (GSE102114; www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102114).
Authors' contributions
SP, JL, SW and JZ were responsible for the design, supervision of the study and revision of the manuscript. JL and SW acquired the data and helped perform the analysis with constructive discussions. SP analyzed and interpreted the patient data regarding the HSPN and was a major contributor in writing the manuscript. GY and XD participated in the designing the study, drafting the manuscript and critically revising it for intellectual content. JZ agreed to being accountable for all aspects of the work to ensure that questions associated with the accuracy or integrity of any part of the study are appropriately investigated and resolved. All authors read, discussed, revised and approved the final manuscript.
Ethics statement and consent to participate
The present study was approved by Ethics Committee of the Institutional Ethics Board of the Affiliated Hospital of Liaoning University of Traditional Chinese Medicine (approval no. 2016CS(KT)-002-01). Written informed consent was obtained by the parents of all of the subjects enrolled in the study.
Patient consent for publication
Written informed consent was obtained by the parents of all of the pediatric patients enrolled in the study for publication of any associated data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen JY, Mao JH. Henoch-Schönlein purpura nephritis in children: Incidence, pathogenesis and management. World J Pediatr. 2015;11:29–34. doi: 10.1007/s12519-014-0534-5. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki Y, Ono A, Ohara S, Suzuki Y, Suyama K, Suzuki J, Hosoya M. Henoch-Schönlein purpura nephritis in childhood: Pathogenesis, prognostic factors and treatment. Fukushima J Med Sci. 2013;59:15–26. doi: 10.5387/fms.59.15. [DOI] [PubMed] [Google Scholar]
- 3.Gardner-Medwin JM, Dolezalova P, Cummins C, Southwood TR. Incidence of Henoch-Schönlein purpura, Kawasaki disease, and rare vasculitides in children of different ethnic origins. Lancet. 2002;360:1197–1202. doi: 10.1016/S0140-6736(02)11279-7. [DOI] [PubMed] [Google Scholar]
- 4.Yang YH, Hung CF, Hsu CR, Wang LC, Chuang YH, Lin YT, Chiang BL. A nationwide survey on epidemiological characteristics of childhood Henoch-Schönlein purpura in Taiwan. Rheumatology (Oxford) 2005;44:618–622. doi: 10.1093/rheumatology/keh544. [DOI] [PubMed] [Google Scholar]
- 5.Saulsbury FT. Clinical update: Henoch-Schönlein purpura. Lancet. 2007;369:976–978. doi: 10.1016/S0140-6736(07)60474-7. [DOI] [PubMed] [Google Scholar]
- 6.Sano H, Izumida M, Shimizu H, Ogawa Y. Risk factors of renal involvement and significant proteinuria in Henoch-Schönlein purpura. Eur J Pediatr. 2002;161:196–201. doi: 10.1007/s00431-002-0922-z. [DOI] [PubMed] [Google Scholar]
- 7.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends Genet. 2014;30:439–452. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu TM, Palanisamy K, Sun KT, Day YJ, Shu KH, Wang IK, Shyu WC, Chen P, Chen YL, Li CY. RANTES mediates kidney ischemia reperfusion injury through a possible role of HIF-1α and LncRNA PRINS. Sci Rep. 2016;6:18424. doi: 10.1038/srep18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Zhang X, Xue C, Zhang H, Shashaty MG, Gosai SJ, Meyer N, Grazioli A, Hinkle C, Caughey J, et al. The long noncoding RNA landscape in hypoxic and inflammatory renal epithelial injury. Am J Physiol Renal Physiol. 2015;309:F901–F913. doi: 10.1152/ajprenal.00290.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One. 2011;6:e18671. doi: 10.1371/journal.pone.0018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan LJ, Ding M, Hou LJ, Cui YT, Li CJ, Yu DM. Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARγ in diabetic nephropathy. Biochem Biophys Res Commun. 2017;484:598–604. doi: 10.1016/j.bbrc.2017.01.145. [DOI] [PubMed] [Google Scholar]
- 13.Huang YS, Hsieh HY, Shih HM, Sytwu HK, Wu CC. Urinary Xist is a potential biomarker for membranous nephropathy. Biochem Biophys Res Commun. 2014;452:415–421. doi: 10.1016/j.bbrc.2014.08.077. [DOI] [PubMed] [Google Scholar]
- 14.Arvaniti E, Moulos P, Vakrakou A, Chatziantoniou C, Chadjichristos C, Kavvadas P, Charonis A, Politis PK. Whole-transcriptome analysis of UUO mouse model of renal fibrosis reveals new molecular players in kidney diseases. Sci Rep. 2016;6:26235. doi: 10.1038/srep26235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B, Xia S, Chen S, Tang Y, Shen N. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res Ther. 2015;17:131. doi: 10.1186/s13075-015-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao X, Duan J, Yan Y, Ma X, Zhang Y, Wang H, Ni D, Wu S, Peng C, Fan Y, et al. Upregulation of long noncoding RNA PVT1 predicts unfavorable prognosis in patients with clear cell renal cell carcinoma. Cancer Biomark. 2017;21:55–63. doi: 10.3233/CBM-170251. [DOI] [PubMed] [Google Scholar]
- 17.Leti F, DiStefano JK. Long noncoding RNAs as diagnostic and therapeutic targets in type 2 diabetes and related complications. Genes (Basel) 2017;8(pii):E207. doi: 10.3390/genes8080207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subspecialty Group of Renal Disease, Society of Pediatrics, Chinese Medical Association: Clinical classification, diagnosis and treatment of glomerular diseases in children. Chin J Pediatr. 2001;39:746–749. [Google Scholar]
- 19.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Liu GL, Gao YF, Xia ZK, Fan ZM, Ren XG, Mao S, He X, Sun T. The pathological analysis of 2551 children patients with glomerulopathy. J Med Postgrad. 2011;24:294–297. (In Chinese) [Google Scholar]
- 22.Hennies I, Gimpel C, Gellermann J, Möller K, Mayer B, Dittrich K, Büscher AK, Hansen M8, Aulbert W, Wühl E, et al. Presentation of pediatric Henoch-Schönlein purpura nephritis changes with age and renal histology depends on biopsy timing. Pediatr Nephrol. 2018;33:277–286. doi: 10.1007/s00467-017-3794-1. [DOI] [PubMed] [Google Scholar]
- 23.Chan H, Tang YL, Lv XH, Zhang GF, Wang M, Yang HP, Li Q. Risk factors associated with renal involvement in childhood Henoch-Schönlein purpura: A meta-analysis. PLoS One. 2016;11:e0167346. doi: 10.1371/journal.pone.0167346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogdanović R. Henoch-Schönlein purpura nephritis in children: Risk factors, prevention and treatment. Acta Paediatr. 2009;98:1882–1889. doi: 10.1111/j.1651-2227.2009.01445.x. [DOI] [PubMed] [Google Scholar]
- 25.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, Lam WL. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li SY, Susztak K. The long noncoding RNA Tug1 connects metabolic changes with kidney disease in podocytes. J Clin Invest. 2016;126:4072–4075. doi: 10.1172/JCI90828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One. 2014;9:e107016. doi: 10.1371/journal.pone.0107016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi HS, Yang XY, Yuan JH, Yang F, Xu D, Guo YJ, Zhang L, Zhou CC, Wang F, Sun SH. H19 inhibits RNA polymerase II-mediated transcription by disrupting the hnRNP U-actin complex. Biochim Biophys Acta. 2013;1830:4899–4906. doi: 10.1016/j.bbagen.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. doi: 10.1186/1479-5876-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu A, Wang Y, Yin J, Zhang J, Cao S, Cao J, Shen Y. Microarray analysis of long non-coding RNA expression profiles in monocytic myeloid-derived suppressor cells in Echinococcus granulosus-infected mice. Parasit Vectors. 2018;11:327. doi: 10.1186/s13071-018-2905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang Y, Zhang Y, Tang Y, Li Q. MALAT1 modulates TGF-β1-induced endothelial-to-mesenchymal transition through downregulation of miR-145. Cell Physilo Biochem. 2017;42:357–372. doi: 10.1159/000477479. [DOI] [PubMed] [Google Scholar]
- 35.Zsóri KS, Muszbek L, Csiki Z, Shemirani AH. Validation of reference genes for the determination of platelet transcript level in healthy individuals and in patients with the history of myocardial infarction. Int J Mol Sci. 2013;14:3456–3466. doi: 10.3390/ijms14023456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozmus CE, Potočnik U. Reference genes for real-time qPCR in leukocytes from asthmatic patients before and after anti-asthma treatment. Gene. 2015;570:71–77. doi: 10.1016/j.gene.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Ding L, Sandford AJ. Selection of regerence genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol Biol. 2005;6:4. doi: 10.1186/1471-2199-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Wang Y, Wang H, Hao X, Wu Y, Guo J. Selection of reference genes for gene expression studies in porcine whole blood and peripheral blood mononuclear cells under polyinosinic: Polycytidylic acid stimulation. Asian-Australas J Anim Sci. 2014;27:471–478. doi: 10.5713/ajas.2013.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SJ, Suh JS, Lee JH, Lee JW, Kim SH, Han KH, Shin JI. Advances in our understanding of the pathogenesis of Henoch-Schönlein purpura and the implications for improving its diagnosis. Expert Rev Clin Immunol. 2013;9:1223–1238. doi: 10.1586/1744666X.2013.850028. [DOI] [PubMed] [Google Scholar]
- 40.Davin JC. Henoch-Schonlein purpura nephritis: Pathophysiology, treatment, and future strategy. Clin J Am Soc Nephrol. 2011;6:679–689. doi: 10.2215/CJN.06710810. [DOI] [PubMed] [Google Scholar]
- 41.Shin JI, Koh H, Lee JS. Henoch-Schönlein purpura associated with helicobacter pylori infection: The pathogenic roles of IgA, C3, and cryoglobulins? Pediatr Dermatol. 2009;26:768–769. doi: 10.1111/j.1525-1470.2009.01039.x. [DOI] [PubMed] [Google Scholar]
- 42.Hirayama K, Kobayashi M, Muro K, Yoh K, Yamagata K, Koyama A. Specific T-cell receptor usage with cytokinemia in Henoch-Schönlein purpura nephritis associated with Staphylococcus aureus infection. J Intern Med. 2001;249:289–295. doi: 10.1046/j.1365-2796.2001.00815.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen T, Guo ZP, Li MM, Li JY, Jiao XY, Zhang YH, Liu HJ. Tumour necrosis factor-like weak inducer of apoptosis (TWEAK), an important mediator of endothelial inflammation, is associated with the pathogenesis of Henoch-Schonlein purpura. Clin Exp Immunol. 2011;166:64–71. doi: 10.1111/j.1365-2249.2011.04442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haslett C. Granulocyte apoptosis and inflammatory disease. Br Med Bull. 1997;53:669–683. doi: 10.1093/oxfordjournals.bmb.a011638. [DOI] [PubMed] [Google Scholar]
- 45.Ozaltin F, Besbas N, Uckan D, Tuncer M, Topaloglu R, Ozen S, Saatci U, Bakkaloglu A. The role of apoptosis in childhood Henoch-Schonlein purpura. Clin Rheumatol. 2003;22:265–267. doi: 10.1007/s10067-003-0718-1. [DOI] [PubMed] [Google Scholar]
- 46.Yuan P, Bo Y, Ming G, Wen-Jun F, Qin Z, Bo H. Apoptosis of human umbilical vein endothelial cells (HUVEC) induced by IgA1 isolated from Henoch-Schonlein purpura children. Asian Pac J Allergy Immunol. 2014;32:34–38. doi: 10.12932/AP0357.32.1.2014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the present study are available from the Gene Expression Omnibus (GSE102114; www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102114).