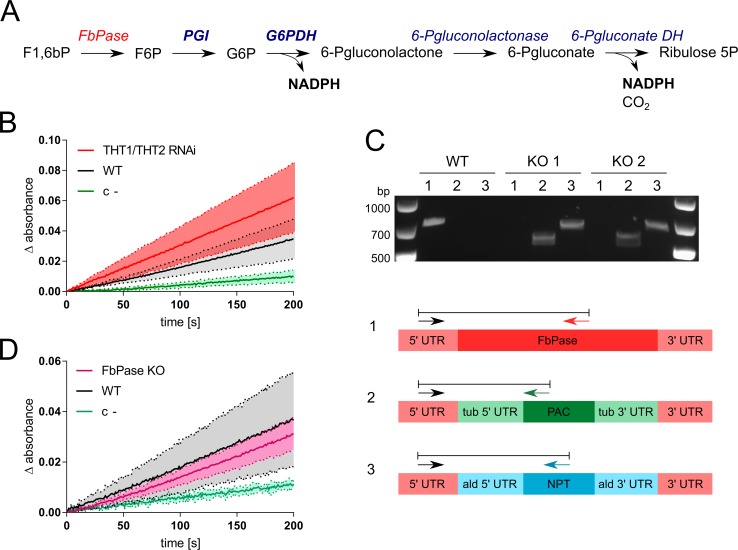
Fig 6. Fructose-1,6-bisphosphatase activity in bloodstream form trypanosomes.
(A) A scheme showing the coupled enzymatic assay used for fructose-1,6-bisphosphatase (FbPase) activity detection. F1,6bP is added to cell extract, and if FbPase is present, it converts F1,6bP into F6P. F6P is converted into G6P by glucose-6-phosphate isomerase (PGI) and further into 6-phosphogluconolactone by glucose-6-phosphate dehydrogenase (G6PDH) producing NADPH. Further, 6-phosphogluconate is made by 6-phosphogluconolactonase, followed by a 6-phosphogluconate dehydrogenase reaction resulting in ribulose 5-phosphate and additional NADPH. PGI and G6PDH were added to the reaction, while the latter two enzymes are present in the cell extract. NADPH production is detected spectrophotometrically. (B) FbPase activity detected in WT (no glycerol in growth medium) and THT1/THT2 knockdown cells induced with tetracycline (plus 5 mM glycerol) for 3 days, and a negative control where F1,6bP was omitted. The lines show means of replicates (n = 3) and shaded areas indicate SD. The slopes measured for WT and THT1/THT2 knockdown cells are significantly different (p < 0.0001, linear regression). (C) Validation of the FbPase (Tb927.9.8720) knockout (KO); two independent populations. The gel shows the PCR assays and the schematic maps indicate the native FbPase gene (lane 1), and alleles after precise replacement with a PAC (lane 2) or NPT cassette (lane 3). (D) FbPase activity detected in FbPase KO cells. The lines show means of replicates (n = 2) for two independent FbPase KO populations. Other details as in A-B above.