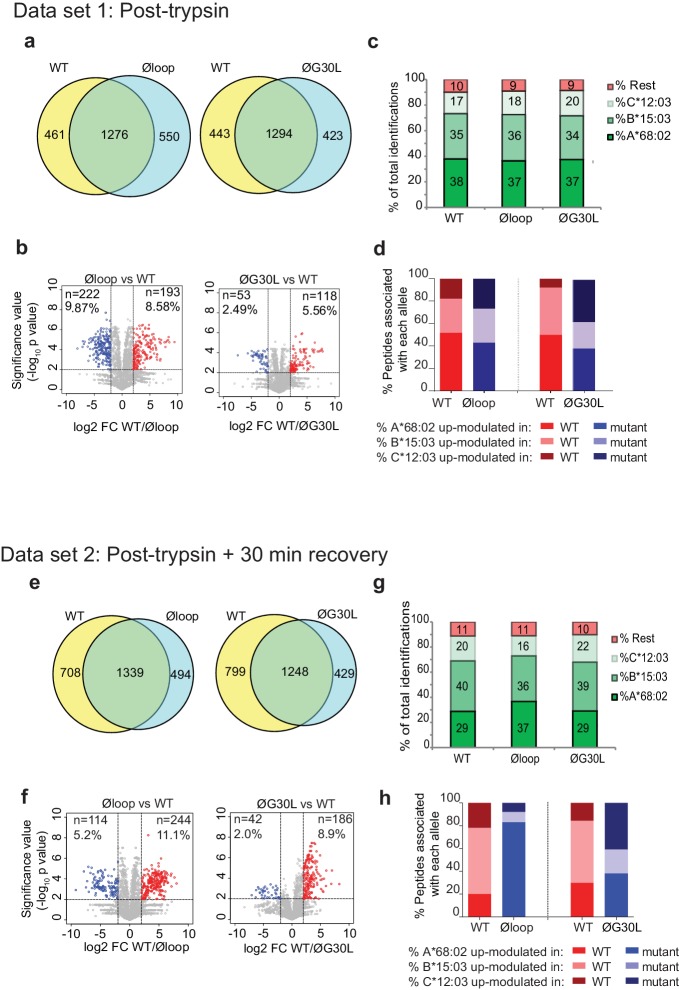
Figure 5. Mutation of the K22-D35 loop of TAPBPR changes the peptide repertoire presented on cells.
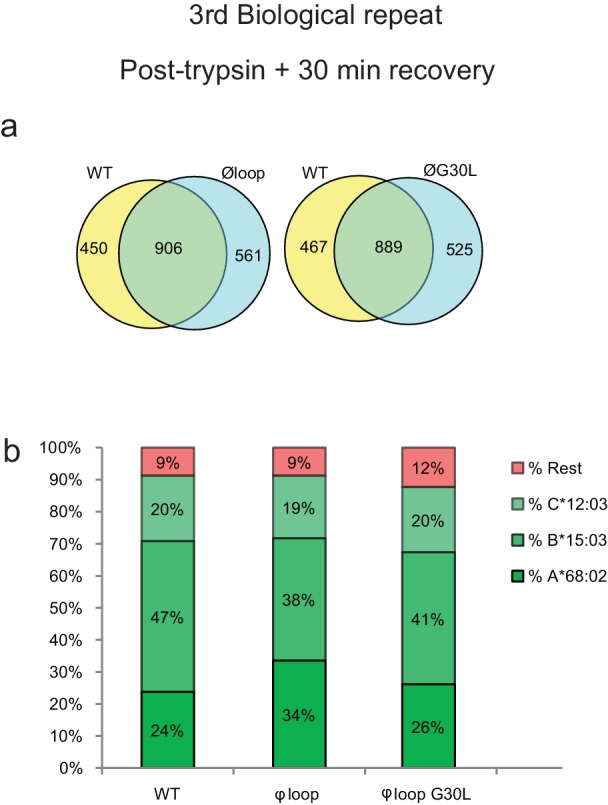
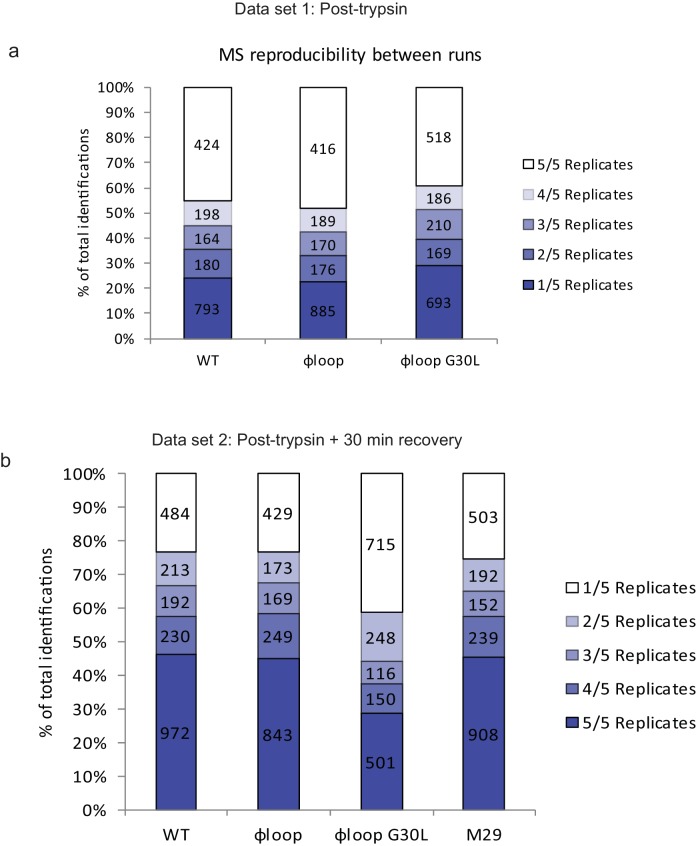
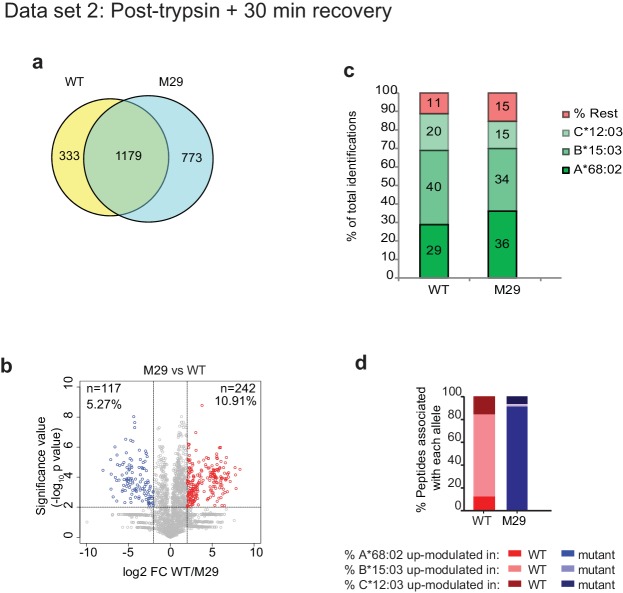
Peptides eluted from W6/32-reactive MHC I complex isolated from IFNγ treated HeLaM-TAPBPRKO expressing either TAPBPRWT, TAPBPRØloop or TAPBPRØG30L were analysed using LC-MS/MS. In dataset 1 (a–d), cells were frozen immediately post-trypsination while in dataset 2 (e–h) cells were allowed to recover in media for 30 min after trypsination, before freezing. The sequences of identified peptides are listed in Figure 5—source datas 2–7. The comparison of all five technical replicates for the two datasets is shown in Figure 5—figure supplement 1. (a,e) Venn diagrams compare all the identified peptides using a presence/absence approach. (b,f) Volcano plots graphically summarise label-free quantitation, displaying modulated peptides between two cells lines. Colour circles highlight the peptide which are differentially expressed between two cell lines after applying an adjusted p-value of <0.01. The list of these peptides is available in Figure 5—source datas 8 and 9. n = number of significantly modulated peptides, % demonstrates the fraction of significantly modulated peptides in a specific cell line compare to all peptides in the comparison. (c,d,g,h) Bar graphs summarise the MHC I molecules (HLA-A*68:02, -B*15:03 or –C*12:03) that the (c,g) identified peptides in a/e, and (d,h) the significantly modulated peptides identified in b/f were matched to using the NetMHCpan-4.0. In (c) and (g), peptides not successfully assigned are indicated in orange (rest). Analysis of the peptide repertoire from a further TAPBPR-loop mutant lacking L30 and from a third biological repeat can be found in Figure 5—figure supplements 2 and 3 respectively. Analysis of the predicted affinity of peptides differential modulated upon mutation of the loop (i.e those in b) and (f) can be found in Figure 5—figure supplement 4.
Figure 5—figure supplement 1. Technical reproducibility of LC-MS/MS measurement.
Figure 5—figure supplement 2. Mutation of residue A29-D35 in the loop of TAPBPR changes the peptide repertoire presented on cells.
Figure 5—figure supplement 3. 3rd biological repeat.