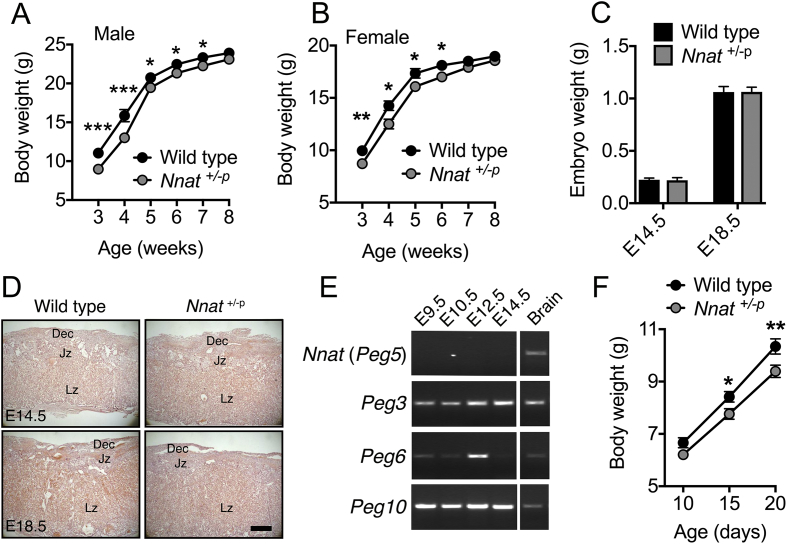
Figure 1.
Juvenile Nnat deficient mice on a 129S2/Sv background display a ‘catch up’ growth phenotype. (A, B) Body weights of male (A) and female (B) wild type and Nnat+/−p (paternal deletion) mice from weaning age (3 weeks) (n = 19 and 17 for wild type and Nnat+/−p mice respectively, n = 17 and 18 for females). Data is represented as mean ± SEM. (C) Wet weights of wild type and Nnat+/−p embryos at E14.5 and E18.5 timepoints (n = 22 and 26 for E14.5 and E18.5 respectively per genotype). Data represented as mean ± SD. (D) Representative H&E-stained sections through placenta taken from wild type and Nnat+/−p embryos at E14.5 and E18.5 (n = 5 mice per genotype). Scale bar = 500 μm. Dec = maternal decidua, Jz = junctional zone, Lz = labyrinth. (E) PCR analysis of Nnat (Peg5), Peg3, Peg6 and Peg10 expression in pooled cDNA extracted from at least four wild type 129S2/Sv placentas between embryonic stages E9.5–14.5. Amplification in pooled cDNA from four 129S2/Sv P0 whole brains served as a positive control. (F) Body weights of wild type and Nnat+/−p male mice from 10 days old (P10) until weaning age (P20) (n = 17 and 20 mice for wild type and Nnat+/−p mice respectively). Data represented as mean ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001).