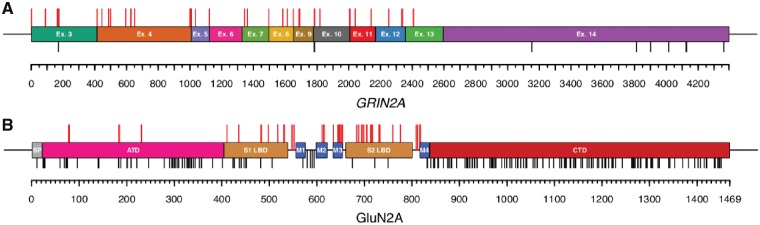
Figure 1.
Distribution of variants. (A) Pathogenic or likely pathogenic null variants (red bars) are spread over nearly the entire gene. However, according to ACMG criteria, the last exon 14 is spared, which encodes nearly the complete C-terminal domain. Null variants in healthy gnomAD controls (black bars) occur primarily in the last exon 14 (probability loss-of-function intolerance 1.00 in ExAC). (B) Pathogenic or likely pathogenic missense variants (red bars) cluster in regions of GRIN2A encoding functionally important domains (S1 and S2 ligand binding domains as well as M1–M4 transmembrane domains and linker regions). The density of missense variants in healthy gnomAD controls (MAC = 2, black bars) is highest in the intracellular C-terminal domain.