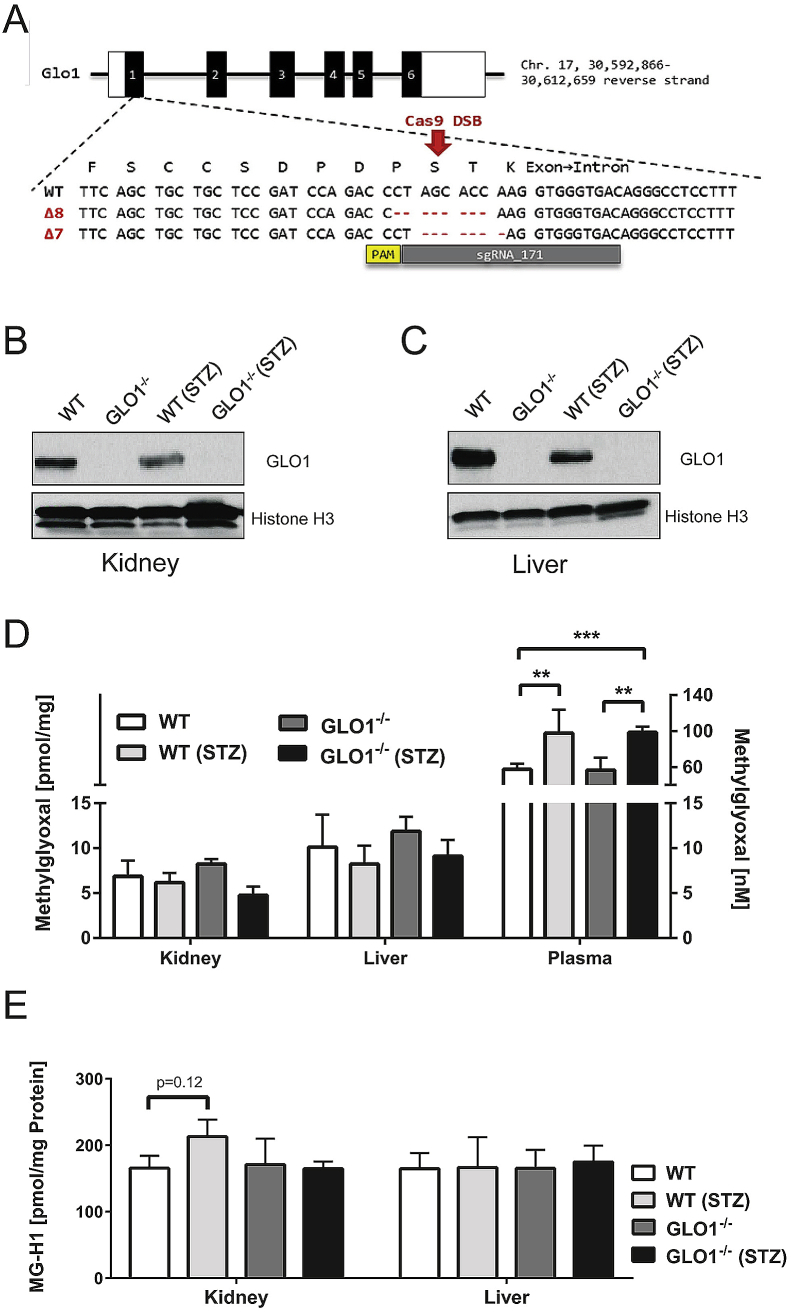
Figure 1.
Schematic overview of CRISPR/Cas9 genome editing in Glo1 locus and T7 assay of found tail DNA & quantitation of GLO1, MG and MG-H1 levels. (A)Glo1 sgRNA target locus in exon 1. The sgRNA binding sequence (sgRNA_171) is depicted in grey and is located on the reverse strand, the corresponding PAM sequence is marked in yellow. Below the WT sequence, the CRISPR/Cas9-mediated deletion of 8 and 7 base pairs leading to a frame shift in the respective Δ8 and Δ7 Glo1−/− mice mouse lines is are shown. B-C: Representative western blotting analysis of total cell extracts from kidney (B) and liver (C) of the appropriate mouse group probed with anti-GLO1 antibody and anti-Histone H3 antibody as a loading control. D: Intracellular MG-levels in whole organs of the appropriate group. E: Intracellular levels of MG-modified arginine (MG-H1) residues after exhaustive enzymatic digestion and determination via LC-MS/MS. Data represent mean ± S.E; *p < 0.05, **p < 0.01, ***p < 0.001 (n = 4).