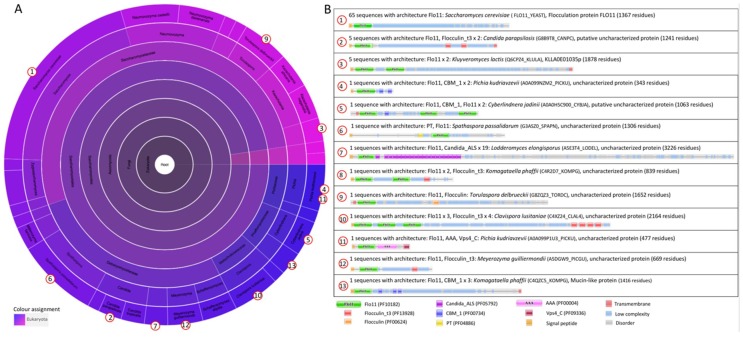
Figure 2.
The family Flo11 (PF10182). (A) Sunburst phylogenetic representation of the Flo11 domain family. (B) Currently (July 2018), the Flo11 domain is present in 13 architectures and only within the ascomycetal orders of the Saccharomycetales. Indicated domains are Flo11 (PF10182); the “Flocculin type 3 repeat” (Flocculin_t3, PF13928) that is found in Flo9 close to its C-terminus and in a number of other Saccharomyces proteins [1]; the “Flocculin repeat” (Flocculin, PF00624) that is rich in serine and threonine residues [2]; “Candida agglutinin-like (ALS)” (Candida_ALS, PF05792) [3,4]; the “carbohydrate-binding module” (CBM_1, PF00734), which is found in carbohydrate-active enzymes [5,6]; the “PT repeat” (PT, PF04886), which is composed on the tetrapeptide XPTX; the ATPase family that is associated with various cellular activities (AAA, PF00004), in which AAA family proteins often perform chaperone-like functions that assist in the assembly, operation, or disassembly of protein complexes [7,8,9]; the “Vps4 C terminal oligomerization” domain (Vps4_C, PF09336) that is found at the C-terminal of ATPase proteins involved in vacuolar sorting, forms an α-helix structure, and is required for oligomerization [41]. The graphics were generated with Pfam version 31.0 [38].