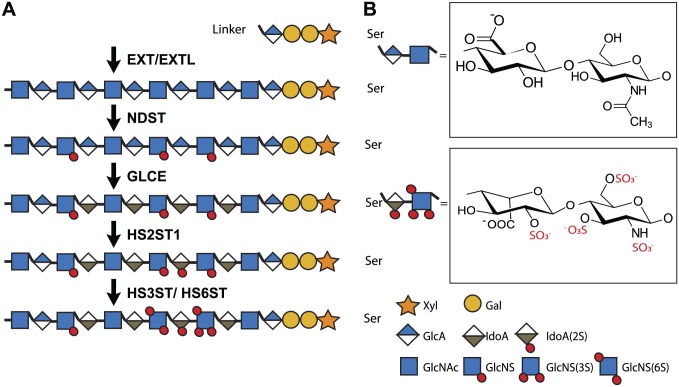
Figure 2.
Enzymes responsible for HS biosynthesis. HS biosynthesis is initiated in the Golgi by the formation of a tetrasaccharide linker region (on serine residues of the core protein) consisting of a xylose, 2 galactose, and a glucuronic acid (GlcA) residue. Subsequently, HS chains are synthesized and modified in a template-independent manner by up to 25 different enzymes. First, the exostosis (EXT) and exostosin-like (EXTL) glycosyltransferase enzyme family elongate the HS chain by alternating the addition of N-acetyl glucosamine (GlcNAc) and GlcA. Next, the N-deacetylase/N-sulfotransferase (NDST) family starts the modification of the HS backbone by deacetylating GlcNAc, followed by N-sulfation. Afterwards, glucuronic acid epimerase (GLCE) converts some GlcA residues into iduronic acid (IdoA) and a 2-O-sulfotransferase (HS2ST1) can then sulfate IdoA. Alternatively, glucosamine can be sulfated by 6-O-sulfotransferases (HS6ST1-3) and 3-O-sulfotransferases (HS3ST1-6). As these enzymes do not completely sulfate the HS chain, they generate complex sulfation patterns that form structurally diverse protein-binding sites. 6- and 3-O-sulfotransferases are thought to be particularly important, since these 10 enzymes have slightly different substrate specificities and are highly evolutionarily conserved. Even after HS biosynthesis in the Golgi system, HS can be extracellularly modified by the endosulfatases sulfatase (SULF) 1 and 2, which can specifically remove sulfate groups from the 6-O position. Moreover, heparanase can degrade the HS chain into shorter oligosaccharides. Together, these biosynthetic enzymes account for the structural diversity of HS.37,38 Abbreviation: HS, heparan sulfate.