Abstract
Almost all eukaryotic mRNAs acquire a poly(A) tail at the 3′-end by a concerted RNA processing event: cleavage and polyadenylation. The canonical PAP, PAPα, was considered the only nuclear PAP involved in general polyadenylation of mRNAs. A phosphoinositide-modulated nuclear PAP, Star-PAP, was then reported to regulate a select set of mRNAs in the cell. In addition, several non-canonical PAPs have been identified with diverse cellular functions. Further, canonical PAP itself exists in multiple isoforms thus illustrating the diversity of PAPs. In this review, we compare two nuclear PAPs, Star-PAP and PAPα with a general overview of PAP diversity in the cell. Emerging evidence suggests distinct niches of target pre-mRNAs for the two PAPs and that modulation of these PAPs regulates distinct cellular functions.
Keywords: Poly(A) polymerase (PAP); Canonical PAP; PAP isoforms; PAPα; Star-PAP; PI4,5P2; PIPKIα; CKI; Polyadenylation; Uridylation; 3′-end processing; Oxidative stress
1. Introduction
In eukaryotes, nuclear mRNA synthesis is a multistep process that begins with transcription and ends with processing at the 3′-UTR [1–5]. The various steps of mRNA synthesis – transcription, splicing, and 3′-end formation are functionally interconnected through a network of synergistic interactions [3,6]. The 3′-end processing of a precursor mRNA (pre-mRNA) is an essential step in eukaryotic gene expression, which is comprised of two steps – cleavage and addition of poly(A) tail [1,4,5,7,8]. Almost all eukaryotic mRNAs are polyadenylated, a step critical for stability, export and translation efficiency of mRNAs [1,4,5,7,9,10]. Pre-mRNAs are polyadenylated by enzymes called poly(A) polymerases (PAPs) which function in a 3′-end processing complex comprised of a large number of protein constituents [11].
The canonical PAP exists in multiple isoforms and at least three forms of canonical PAP – PAPα, PAPβ (PAPT), and PAPγ (neoPAP) have been reported [12–18]. Canonical PAPs were considered the only PAPs that controlled all co-transcriptional polyadenylation in the nucleus. Apart from PAPα, PAPγ also functions in the similar CPSF and AAUAAA signal dependent polyadenylation of pre-mRNAs in the nucleus [13]. Another nuclear non-canonical PAP, Star-PAP [Speckle Targeted PIPKIα Regulated Poly(A) Polymerase] (RBM21, TUT1), was then reported to polyadenylate certain mRNAs involved in various cellular processes such as oxidative stress response and apoptosis [19,20]. Based on similarities in domain architecture, Star-PAP belongs to a subfamily of non-canonical PAPs (ncPAPs). So far, seven known ncPAPs have been reported in humans with diverse cellular functions [21,22].
PAPα and Star-PAP participate in both cleavage and polyadenyalation reactions. In addition, Star-PAP exhibits terminal uridylyl transferase activity toward U6 snRNA [23]. Do the two PAPs compete for target mRNAs? Emerging evidence suggests distinct niches of target mRNAs for each PAP and there appears to be no cross regulation of their targets [19,20,24]. Such specificities in target poly(A) site recognition could potentially modulate alternative polyadenylation (APA). This review presents an overview of diverse PAPs in the cell and compares two functionally similar but distinct nuclear PAPs, Star-PAP and PAPα. Here, we explore the differences in properties, mechanism, target mRNA selection, and regulation of the two PAPs in 3′-end pre-mRNA processing.
2. 3′-end pre-mRNA processing in gene expression
The 3′-UTR of an mRNA is critical for the regulation of gene expression. During eukaryotic mRNA maturation, the nascent pre-mRNA undergoes processing at the 3′-UTR. This processing at the 3′-end is a two-step event: first, the pre-mRNA is endonucleolytically cleaved at the cleavage site, followed by the addition of poly(A) tail to the upstream fragment of the cleaved RNA, while the downstream fragment is rapidly degraded [1,4,5,7,8,10]. 3′-end processing is intricately coupled to transcription and splicing, and also regulates the type, and the amount of mRNA and protein levels of a particular gene. Thus, mRNA 3′-end formation links transcription of a gene with the translation of its mRNA [2,6].
Mass spectrometry analysis identified ~85 protein factors associated in the 3′-end processing complex [11]. Some of the critical proteins required for the cleavage and polyadenylation reactions include subunits of Cleavage and Polyadenylation Stimulatory Factor (CPSF), Cleavage stimulatory Factor (CstF), Cleavage Factor Im (CF Im), and Cleavage Factor IIm; Symplekin, PAP, and the nuclear Poly(A) Binding Protein (PABPN1) (for review [5,25]). Mammalian CPSF consists of six polypeptides – CPSF 160 (CPSF1), CPSF 100 (CPSF2), CPSF 73 (CPSF3), CPSF 30 (CPSF4), hFip1 and WDR33. CPSF 160 recognises the poly(A) signal, PAS (AAUAAA), a sequence located approximately 15–30 nucleotide upstream of cleavage site and interacts with PAP and CstF [26]. Although CPSF 160 binds to the AAUAAA signal and cooperates with other factors, the assembly of the stable cleavage complex requires an intact CPSF complex [26–28]. The CPSF interaction also requires cooperation with CstF and CF Im for stable association with pre-mRNA [29–32]. Studies suggest that other trans-acting factors such as splicing factor U1 snRNP interacts with CPSF 160 and promotes its binding to PAS on the pre-mRNA [33]. In HIV, CPSF 160 can also interact with sequence element upstream of the poly(A) site other than the classical AAUAAA signal at the 3′-UTR [34,35]. Another subunit of CPSF, CPSF 73, acts as endonuclease, binds directly to the cleavage site in a AAUAAA dependent manner and then cleaves the pre-mRNA at the cleavage site [36,37]. CPSF 30 may cooperate with CPSF 160 in RNA binding [38]. hFip1, one additional CPSF subunit, also binds PAP and directs PAP to the cleavage site [39]. The exact functions of CPSF 100 and WDR33 subunits are yet undefined [11,40].
The CstF complex recognises the GU/U rich downstream sequence element (DSE) and cooperates with CPSF. CstF has three subunits – 50 (CSTF1), 64 (CSTF2) and 77 (CSTF3) KDa of which CstF 64 binds the GU/U rich downstream sequence element (DSE) [30,41,42]. CstF 77 functions as a homodimer and bridges the 64 and 50 KDa subunits and cooperates with CPSF 160; CstF 50 interacts with the RNA Polymerase II (Pol II) C-terminal domain (CTD) [26,43,44]. The interaction of CstF and CPSF complexes and their corresponding associations with DSE and PAS is considered the most significant event in defining the cleavage site. CF Im is a heterotetramer with two 25 KDa subunits (CPSF5 or NUDT21) that forms the core of the complex along with two larger polypeptides of 68 KDa (CPSF6) and/or 59 KDa (CPSF7) subunits. CPSF5 binds pre-mRNA upstream of PAS to a sequence element that contains the U(G/A)UA motif. In addition, CF Im cooperates with CPSF for RNA binding and enhances the recognition of the cleavage site [31,45–47] CF Im can also direct a sequence-specific AAUAAA-independent polyadenylation by recruiting the CPSF subunit hFip1 and PAP in vitro [48]. CF IIm consists of two subunits, hPcfl 1 and hClp1, and possibly links CF Im and CPSF within the cleavage complex [49]. Symplekin is a scaffolding protein that putatively joins a large number of proteins together in the complex [50–53]. CPSF, CstF, Symplekin and CF Im interact with each other stabilising the 3′-end processing complex assembled on pre-mRNA and promotes recruitment of PAPα. Mammalian PAPα is also required for the cleavage reaction, however, the mechanism as to how PAPα is involved in cleavage is not precisely defined [5,25]. After cleavage of the transcript, PAPα adds a poly(A) tail to the upstream fragment of cleaved RNA. The nuclear poly(A) binding protein (PABPN1) binds the nascent poly(A) tail, confers processivity to PAP and controls poly(A) tail length. PABPN1 also interacts with PAPα and CF Im and enhances the efficiency of polyadenylation [54–60]. Thus, a large number of protein factors cooperate with each other and assemble at the 3′-UTR to accomplish cleavage of the transcript followed by polyadenylation.
3. Polyadenylation
Polyadenylation is a process of template-independent addition of a long poly(A) tail to the 3′-end of an mRNA. Polyadenylation activity was first identified some 50 years ago from calf thymus nuclei extracts [61]. However, it was only a decade later that poly(A) tails were recognised as a product of post-transcriptional processing of the mRNA 3′-UTR [62–64]. Almost all mammalian mRNAs have a poly(A) tail at their 3′-end, with the exception of histone mRNA which ends after a highly conserved RNA stem-loop structure, and lacks a poly(A) tail [65]. The length of a nascent polyadenylated tail on an mRNA in mammalian cells varies from 200 to 300 adenosine residues [1,9]. In the nucleus, the poly(A) binding protein, PABPN1 helps to define the length of the newly synthesised poly(A) tail during de novo mRNA synthesis [9,54,56]. PABPN1 interacts with the first 11 polyadenosine residues added, stimulates PAPs affinity for RNA substrate, and in presence of CPSF induces PAP from its distributive mode to a processive polyadenylation [54,60,66]. When polyadenylation reaches ~250 residues, PAP switches back to its distributive mode resulting effectively in termination of polyadenylation [58]. The precise mechanism of this length control is not fully known, it appears to occur through the formation of a ~20 nm spherical structure involving the poly(A) tail and the bound PABPN1 disrupting the tripartite, CPSF-PAP-PABPN1 processive polyadenylating complex [56,67]. Additionally, a role of multi-functional protein nucleophosmin (NPM1) in poly(A) tail length determination has also been proposed [68,69]. However, a recent poly(A) tail profiling indicated much shorter average lengths of poly(A) tails from various eukaryotic species (<100 in mammalian cells) [70] likely due to the shortening in the cytoplasm [71]. Another genome wide measurement of poly(A) tail length also demonstrated a median tail length of 50–100 adenosine nucleotides in HeLa and NIH 3T3 cells [72]. The observed length of mammalian poly(A) tails is at least influenced or maybe determined by the shortening reaction in the cytoplasm. In addition, there was diversity in the tail length not only among the transcripts from different individual genes but also within different mRNA transcripts from the same gene. Intriguingly, shorter tails were observed for mRNAs encoding ribosomal or housekeeping proteins [70]. In general, poly(A) tails play crucial roles in maintaining mRNA stability and turnover, transport of message from nucleus to cytoplasm, and translation efficiency of mRNA [1,5,9,10]. Moreover, defective polyadenylation has been linked to various human diseases [73].
3.1. Alternative polyadenylation (APA)
In humans, pre-mRNAs are polyadenylated in several different ways due to the existence of more than one polyadenylation site, allowing a single gene to encode multiple mRNA transcripts [74,75]. More than half of the genes in the human genome are alternatively polyadenylated [76]. APA regulation is an important event in the gene expression pathway, critical for a number of diseases [73,75,77–79]. Specific changes in the APA pattern have been observed during cancer progression, stem cell development, and tissue specific expression of genes, yet the mechanism determining particular APA site(s) remains elusive [78,80–90]. APA changes the length of the 3′-UTR thus affecting the miRNA binding sites, or in certain cases the coding region in the mRNA resulting in proteins with different domains [78,81,84,91,92]. Therefore, APA potentially alters the dynamics and properties of a transcript affecting stability, translation and/or subcellular localisation.
4. Poly(A) polymerase (PAP)
PAPs are the enzymes involved in the polymerisation of adenosine residues to form long poly(A) tails at the 3′-end of eukaryotic mRNAs. PAPs are involved not only in the polyadenylation reaction but also in the CPSF mediated cleavage reaction [1]. The gene structure of PAP indicated possible different PAP isoforms by alternative RNA processing [93]. Consistently, different PAP isoforms have been isolated from various sources [18,94–96].
4.1. Canonical PAP (PAPα) – the nuclear enzyme
Canonical PAPα belongs to a nucleotidyl transferase superfamily of DNA Polymerase β [97,98], and is responsible for the polyadenylation of nascent mRNAs in the nucleus. PAPα has three distinct domains: a catalytic domain at the N-terminus, an RNA binding region, and two C-terminal nuclear localisation signals (NLS1 and NLS2) followed by a ~20 KDa extended region enriched in serine (S) and threonine (T) residues [99–102]. Crystal structure of mammalian PAPα showed an aspartate triad in the active site similar to the DNA pol β coordinating the metal ions required for catalysis and ATP recognition [99,101,103,104]. Interestingly, the C-terminal S/T enriched region is highly phosphorylated [99,100,102,105,106]. Moreover, there are seven cyclin dependent phosphorylation sites in this region [106–108]. Phosphorylation at these sites represses the PAP activity. Phosphorylation of PAP will be discussed in a later section. In addition, interaction sites for U1A and U2AF65 splicing factors are present at the C-terminus [109–111] indicating the regulatory potential of the C-terminal end.
4.2. Multiple isoforms of canonical PAP
In humans, three genes encode the canonical PAP – PAPOLA (PAPα), PAPOLB (PAPβ or T) and PAPOLG (PAPγ or neoPAP). While PAPα is ubiquitously expressed, PAPβ is testis-specific and regulates transcripts expressed during spermatogenesis [12,14,15]. Both PAPT and PAPγ share similar structural motifs and sequence identity with PAPα, except for the divergent C-termini. These different forms of PAP (PAPα, β, or γ) are believed to have arisen from a common PAP by gene duplication [12,13,15]. PAPγ appears to have the same function in cleavage and polyadenylation as that of PAPα. Interestingly, PAPγ exhibits monoadenylation activity towards small RNAs in addition to its normal PAP activity [112]. Furthermore, PAPγ was found to be specifically active during tumourigenesis, thus suggesting functional diversity [13,17]. Apart from these different forms of PAP, there exists other PAP related genes in humans. At least two such PAP related gene sequences have been identified from the in silico searches in the human genome [113].
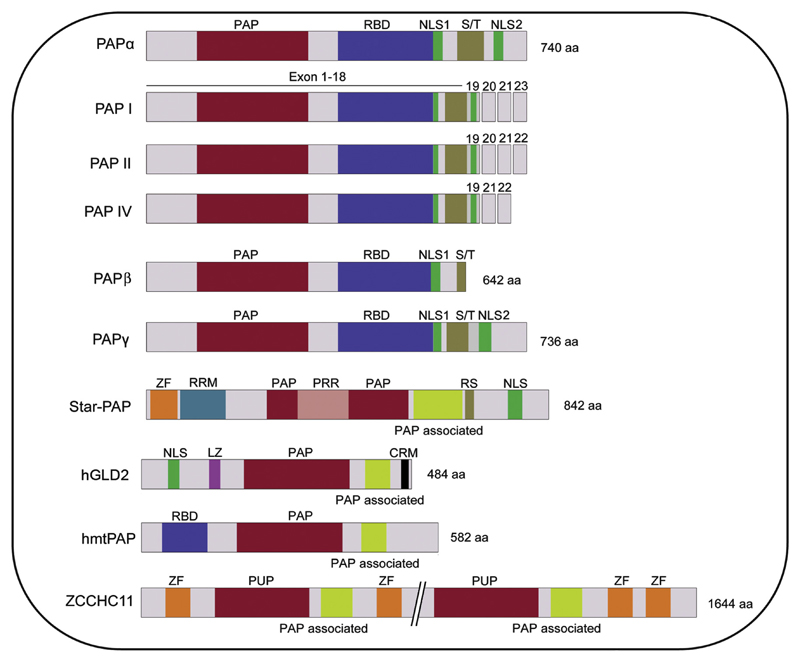
There are at least six isoforms of canonical PAPα generated by alternative splicing, PAP I–VI [16,18,96]. PAPs I, II and IV are longer versions with the full length catalytic domain while III, V and VI are truncated PAPs lacking parts of the catalytic domain. PAPs I, II and IV are functionally active, and are generated by alternative splicing of the last three exons [16,18]. PAP II is the predominant PAP isoform in most cell types [16,94,114]. Truncated PAPs, PAPs III, V and VI lack NLSs, the extended C-terminus in addition to parts of the catalytic domain. These PAPs are generated by alternative polyadenylation and/or splicing events, and do not encode functional proteins in vivo [18,100,115]. Two additional longer PAP isoforms (PAP VIII and IX) generated by alternative splicing of exons 20, 21 and 22 have also been reported [116]. However, at this time the significance of divergent C-termini of the full length PAPs is unclear. Interestingly, the C-terminal S/T rich region which is present in all longer PAP isoforms is dispensable for its activity in vitro [16,102]. Therefore, it is likely to act as a regulatory domain, and it could have a selective advantage of differential interaction with other distinct trans-acting cleavage factors or regulators resulting in functional diversity. Moreover, studies have shown distinct cellular functions for different PAP isoforms in plants [117]. Schematics of various human PAP isoforms have been depicted in Fig. 1.
Fig. 1.
Schematic representation of the domain architecture of human PAPs. Canonicals – PAPα, three functional splice isoforms, PAPβ (PAPT), PAPγ (neoPAP); and non-canonicals – Star-PAP, hGLD2 (PAPD4), hmtPAP (PAPD1), ZCCHCH11 are shown. All canonical PAPs including the splice isoforms have similar structural organisation (except divergence at the C-terminus), comprising a catalytic domain (PAP domain) – red, an RNA binding domain – blue, nuclear localisation signals – green, and a C-terminal Ser/Thr rich regulatory region – brown. Non-canonical PAPs have diverse organisation comprised of a catalytic domain (PAP or PUP) – red, a PAP associated domain – yellow, RNA recognition motifs (such as ZF – orange, Lucine Zipper – magenta, an RNA binding domain – blue, RNA recognition motif – light blue), and Nuclear localisation signals – green. In addition, Star-PAP PAP domain is split by a proline rich region (pink). The size of each protein is indicated (not to scale).
4.3. Non-canonical PAP (ncPAP) – PAPs with functional diversity
ncPAPs are PAP-related members of the Pol β superfamily involved in diverse cellular functions as detailed below. Unlike the canonical counterparts, which add long poly(A) tail during mRNA maturation, ncPAPs typically add short terminal tails and target a variety of substrates (snRNA, miRNA, aberrant rRNA, snoRNA, histone mRNA, etc.). Surprisingly, there are reports of polyadenylation of select pre-mRNAs by one of the ncPAP, Star-PAP (discussed in detail in the following sections) [19,20]. This is an unusual function for an ncPAP as most ncPAPs add short tails. In addition, ncPAPs have distinct domain architecture. For example, all ncPAPs contain a conserved (among ncPAPs) PAP associated domain immediately following the catalytic (PAP) domain (Fig. 1). There are at least seven potential ncPAPs in humans (PAPD1, PAPD4, PAPD5, POLS, RBM21, ZCCHC6, ZCCHC11). PAPD1 (hmtPAP) is a mitochondrial PAP that polyadenylates mitochondrial mRNA [118]. PAPD1 mediated polyadenylation can generate UAA stop codon in some mitochondrial mRNAs, which is not encoded by the mitochondrial DNA [119–121]. However, the functional significance of mitochondrial polyadenylation is still a topic of debate [118,121,122]. PAPD1 was also reported to uridylate histone mRNA along with PAPD5 to target it for degradation [123] although the actual PAP that uridylates histone mRNA is somewhat controversial [124]. hGLD2 (PAPD4) is a cytoplasmic PAP that polyadenylates short (A)-tailed mRNAs in the cytoplasm [125,126] and is involved in diverse functions such as embryonic development, cell cycle, germline maturation, synaptic plasticity, learning and memory [126–131]. hGLD2 also polyadenylates p53 mRNA in the cytoplasm [128,132]. hGLD2 targets mRNAs containing a cytoplasmic polyadenylation element (CPE) at the 3′-UTR that is recognised by the regulatory protein CPEB (CPE binding protein). Phosphorylated CPEB interacts with CPSF 160 and helps recruit the CPSF complex (CPSF 160, 100 and 30) to the PAS. This complex is then stabilised by another processing factor, symplekin. This processing complex then recruits the cytoplasmic PAP, hGLD2 at the 3′-end to elongate the poly(A) tail [129,133,134]. Unlike the nuclear polyadenylation complex, the CPSF 73 subunit is not present in the cytoplasmic polyadenylation complex [135].
PAPD5 and POLS (PAPD7) are two human orthologues of yeast Trf4 involved in nuclear surveillance of a wide range of nuclear target RNAs [136–139]. Both PAPs also polyadenylate aberrant rRNA precursors to target them for degradation [139,140]. PAPD5 has also been implicated in the processing of small nucleolar RNAs (snoRNAs). PAPD5 oligoadenylates late intermediates of H/ACA box snoRNAs during 3′-end shortening, which are then trimmed by the exonuclease PARN to generate mature 3’-ends [141]. Another ncPAP, Star-PAP (RBM21), controls 3′-end processing of mRNAs involved in oxidative stress response [20]. Star-PAP also shows uridylation activity toward U6 snRNA substrate [23] (discussed in detail in subsequent sections). ZCCHC11 (TUT4) and ZCCHC6 (TUT7) are orthologues of yeast Cid1 uridyl transferase with extensive homology to each other and regulate diverse RNA species [21,124,142–145]. ZCCHC11 and ZCCHC6 uridylate miRNA let-7 precursor through interaction with Lin 28, thus controlling let-7 biogenesis [144,146–148]. Further, ZCCHC11 mediated uridylation controls microRNA, miR-26 activity [145]. and has also been implicated in histone mRNA degradation [124]. Thus, the existence of various canonical and non-canonical PAPs suggests diverse functional significance of PAPs in the cell. A list of various human canonical and non-canonical PAPs can be found in Table 1.
Table 1.
List of various human canonical and non-canonical PAPs.
| Name of the PAP | Localisation | Functional significance | Reference |
|---|---|---|---|
| Canonical PAP | |||
| PAPα PAP I PAP II PAP III PAP IV PAP V PAP VI |
Nuclear |
PAP II is the most predominant PAP; involves in general 3′-end processing of all nuclear nascent pre-mRNAs (PAP I and IV – function not clear, likely similar to PAP II) PAP V, III, VI are truncated inactive form, Do not encode functional proteins |
[16,18,94,102,114] |
| PAP β (PAPT) | Nuclear/Cytoplasmic | Testes specific – spermatogenesis | [12,15] |
| PAP γ/neoPAP | Nuclear | Tumourigenesis, monoadenylation activity towards small RNA | [13,17,112] |
| Non-canonical PAP | |||
| PAPD1 (hmtPAP) | Mitochondrial | Mitochondrial mRNA stabilisation, histone mRNA degradation, stop codon regeneration | [118,120–123] |
| PAPD4 (hGld2) | Nuclear/cytoplasmic | Cytoplasmic mRNA polyadenylation, miRNA stabilisation | [122,125,126,128] For review [127,129] |
| PAPD5 | Nuclear | Aberrant rRNA degradation, histone degradation, processing of snoRNAs, various other RNA targets | [21,139–141] |
| POLS (PAPD7) | Nuclear | Not clearly defined, likely redundant to PAPD5 | [21,140] |
| ZCCHC6 (TUT 4) | Nuclear | Similar to ZCCHC11; regulate Let 7 biogenesis | [147,157] |
| ZCCHC11 (TUT 6) | Nuclear | miRNA regulation (let7, mi26a and others), histone mRNA degradation | [124,143,147,157] |
| Star-PAP (RBM 21, TUT1) | Nuclear | Oxidative stress response, DNA damage induced apoptosis and various other cellular functions | [19,20,23,24] |
4.4. Star-PAP – a schizophrenic polymerase
Star-PAP (Speckle Targeted PIPKIα Regulated Poly(A) Polymerase) is a nuclear ncPAP regulated by lipid messenger phosphatidyl inositol 4,5 bisphosphate (PI4,5P2) [20,149–151]. Star-PAP was identified as an interacting partner of phosphatidyl inositol phosphate kinase Iα (PIPKIα) [20]. Star-PAP contains a PAP domain split by a proline rich region (PRR) of ~200 amino acids that is phosphorylated by casein kinase I (CKI) isoforms α and ε [152,153]. Star-PAP has two polynucleotide binding domains – a zinc finger (ZF) and an RNA recognition motif (RRM), both required for Star-PAP RNA binding [24]. Star-PAP and PAPα have similar function but with a distinct mechanism regulating a select set of mRNAs [20,24]. Star-PAP along with PIPKIα specifically control genes involved in the oxidative stress response such as heme oxygenase-1 (HO-1) and NAD(P)H:Quinone Oxidoreductase (NQO-1) [20,152] and the pro-apoptotic gene Bcl-2 interacting killer (BIK) [19].
Star-PAP was initially identified as U6 terminal uridylyl transferase (TUTase), which uridylates U6 snRNA [23] involved in cellular splicing [23,154]. Concomitantly, siRNA knockdown of Star-PAP was reported to dramatically reduce cell viability. However, the exact reason for loss in cell viability is unclear. Given Star-PAP’s apparent role in 3′-terminal modification of U6 snRNA, it is tempting to think of a global impact of Star-PAP on splicing efficiency. However, at present, there is no direct evidence to suggest any role for Star-PAP or a U6-TUTase-catalysed reaction in cellular splicing [23,155,156]. Alternatively, Star-PAP as a PAP is involved in various cellular processes including stress response and apoptosis. Microarray data indicated that a number of Star-PAP targets are genes critical for cell survival [19,20]; thus, loss of Star-PAP could result in cell death. Surprisingly, the study that identified Star-PAP as a PAP did not report any significant effect of siRNA Star-PAP knockdown on the cell viability [20]. In addition, the yeast counterpart Cid11 is not required for cell viability [22]. Nevertheless, Star-PAP uridylation activity has been confirmed in vitro [20,23].
Is Star-PAP a PAP or a TUTase or both in vivo? Although Star-PAP has both PAP and TUTase activities, at the physiological concentration of ATP adenylation activity competes over uridylation suggesting a possible predominance of polyadenylation function in the cell [20]. Nonetheless, with a clear in vitro TUTase activity of Star-PAP, one can envisage a distinct physiological role of Star-PAP uridylation function in the cell. In fact, many ncPAPs have both uridylation and adenylation functions [118,121–123,140]. Therefore, it is conceivable that Star-PAP has a complex role, for example, while polyadenylation regulates a select set of mRNAs, uridylation regulates U6 or other target RNAs. Another possibility is that both activities combined result in a heterogeneous 3′-end tail with both A’s and U’s. A related example has been reported in fission yeast where few Us were incorporated at the end of the 3′-poly(A) tail [157,158]. This modification has been implicated in a new pathway of 5′–3′ mRNA degradation in fission yeast, where addition of short terminal Us at the end of polyadenylated mRNAs appears to stimulate decapping and initiates the mRNA degradation pathway [158]. Widespread uridylation has also been reported downstream of the mammalian mRNA poly(A) tail [72]. Furthermore, Dis3L2, a new 3′–5′ exonuclease, involved degradation of oligouridylated RNAs, was recently identified in multiple eukaryotes [159–161]. Dis3L2 functions independent of the exosome, and uridylation of its targets acts as an RNA decay signal for Dis3L2. Moreover, addition of >10 uridines to the 3′-end of its miRNA target stimulates Dis3L2’s enzymatic activity in vitro [159–161]. These studies also suggest a possible general role of terminal uridylation as signal for RNA decay in eukaryotes. Thus, presence of U’s in the poly(A) tail will have a profound impact on the functional dynamics of an mRNA. Unfortunately, no sequence information of Star-PAP target poly(A) tails is available so far, and hence worthwhile exploring. Such properties, as mentioned above, would make Star-PAP schizophrenic in nature, or a truly non-canonical enzyme.
5. Mechanism of 3′-end RNA processing – Star-PAP versus canonical PAP
Cis-elements present at the 3′-UTR play an important role in specific cleavage and polyadenylation of pre-mRNAs. There are at least four consensus cis-elements at the canonical 3′-UTR [7]. The first important motif is a conserved hexamer AAUAAA (PAS), which determines the position of the cleavage site. Mutation in this consensus hexamer affects the efficiency of 3′-end processing [162,163]. Point mutations in the AAUAAA signal have often been linked to human diseases. For example, two point mutations in the AAUAAA hexamer have been characterised in patients suffering from thalassaemia (AAUAAG in the α2-globin and AACAAA in the β-globin) [164,165]. An A-G point mutation (AAUAAA – AAUGAA) in FOXP3 gene results in a rare X-linked autoimmune disorder, IPEX (immunodysregulation polyendocrinopathy enteropathy X-linked syndrome) [166]. However, variation in the second nucleotide in the consensus hexamer to U (AUUAAA) is the most widely occurring PAS after AAUAAA (for reviews [1,167]). Studies on human and mouse ESTs have indicated AAUAAA (70%) as the most common hexamer followed by AUUAAA (15–20%) among all PAS containing ESTs [76,168]. Analysis of human UTRs and ESTs has indicated alternate signals as well. AGUAAA, UAUAAA, CAUAAA, GAUAAA, AAUAUA, AAUACA, AUAGA, and ACUAAA are few other widely occurring hexamers [168,169]. However, such variants (including the common AUUAAA hexamer) are less efficiently processed than the classical AAUAAA [168]. The second important cis-element at the 3′-UTR is the cleavage site situated ~15–30 bases downstream of PAS. This position of the cleavage site is determined by the locations of both PAS and DSE [30,170]. Although the sequence at the cleavage site is not very conserved, in vertebrates the majority of the cleavage sites are located immediate downstream of an adenosine residue (with preference for A over G), the most optimal site being CA [25,163]. In fact, in the human prothrombin gene, the position of the cleavage site is mutated from weaker CG to most optimum CA, inducing the expression, to cause a mild thrombophilia phenotype [171–173]. The third element is a GU/U-rich DSE located ~20–40 bases downstream of the cleavage site, which also influences the position of the cleavage site. It is less conserved than the PAS and has either a GU-rich (YGUGUUYY, where Y = pyrimidine) or a U-rich (UUUUU) element [174–177]. And, finally the last cis-element is a U-rich, upstream sequence element (USE) situated ~1–20 nucleotides upstream of PAS [7]. USEs can influence the efficiency of PAS and/or complement for the suboptimal PAS or DSE [35,178–181]. In addition, there are auxilary upstream and downstream sequence elements. While the auxiliary upstream sequence mostly contains either U rich (UUUU) or UAUA/UGUA, the auxiliary downstream sequences are generally G-rich [182–185] (for review [5,25]). They are less conserved in position as well as sequence than the four core elements.
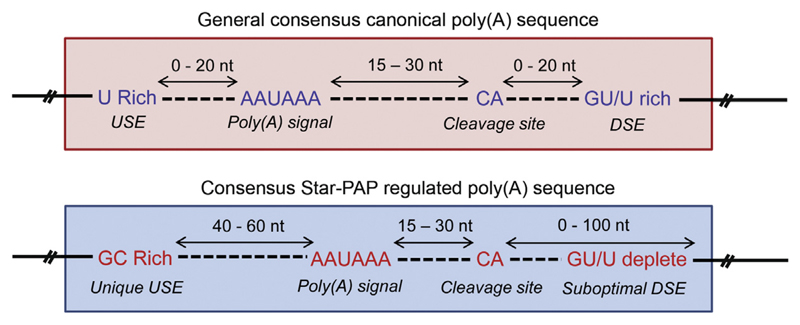
Except for the intact AAUAAA signal, Star-PAP target messages harbour other cis-elements at the 3′-UTR that are distinct from that of canonical 3′-UTR [19]. Star-PAP targets have suboptimal DSEs, with U/GU deplete sequence, which potentially renders CstF dispensable for Star-PAP mediated 3′-end processing. In addition, there is no noticeable canonical motif like USE; instead, a ~40–60 nucleotides long GC-rich sequence of Star-PAP binding region upstream of the PAS is present [19,24]. These variations in the cis-regulatory elements of Star-PAP and canonical PAP target 3′-UTRs explain the mechanistic differences of the two PAPs. A comparison of the 3′-UTR cis-elements of canonical and Star-PAP regulated genes is depicted in Fig. 2.
Fig. 2.
3′-UTR consensus cis-elements of canonical and Star-PAP target 3′-UTRs. The canonical 3′-UTR has distinct consensus elements such as the AAUAAA poly(A) signal, cleavage site ~15–30 nucleotide downstream of PAS, G/GU rich DSE and a U-rich USE (for a review see Ref. [7]). In case of Star-PAP target mRNAs, except for the intact PAS (AAUAAA), they have suboptimal DSEs (deplete U sequence), and no regular USE present; instead, a GC-rich Star-PAP binding region is found around 40–50 nucleotides upstream of the cleavage site is present.
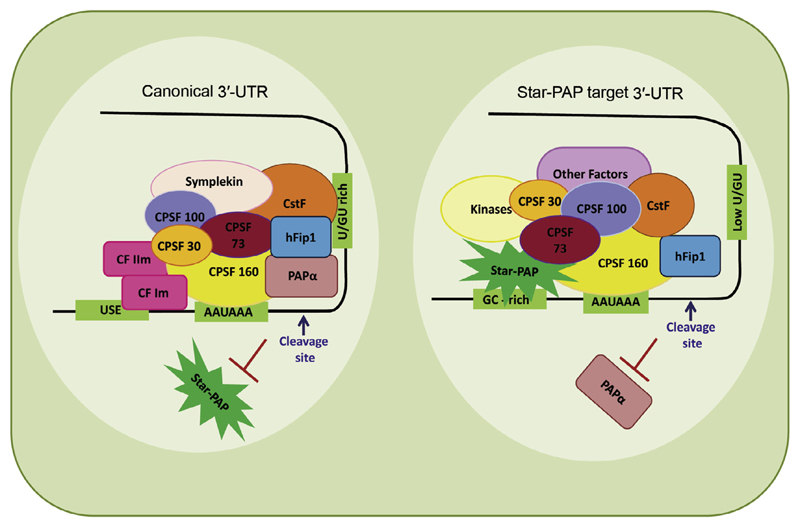
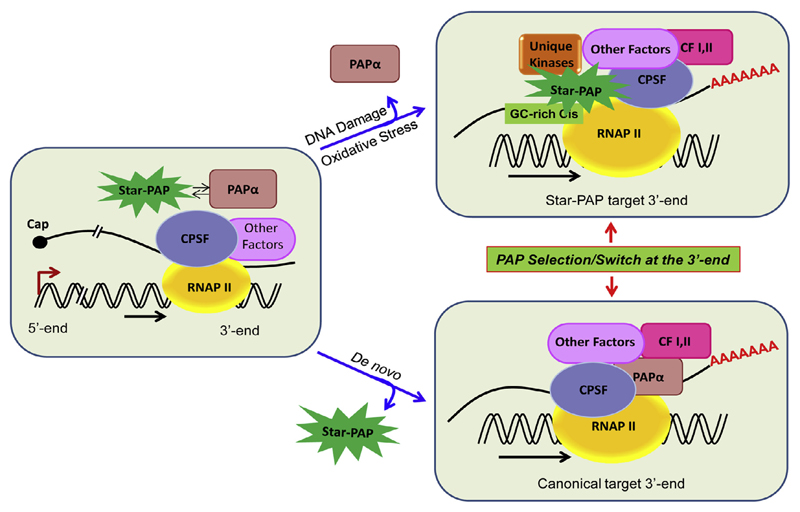
Recent advances in the understanding of 3′-end processing have indicated distinct mechanisms for different PAPs. While PAPα is mechanistically well explored, studies on Star-PAP are still emerging. The two PAPs share similar cleavage factors but assemble distinct 3′-processing complexes and control specific sets of target mRNAs in the cell. Around 85 proteins are associated with the canonical 3′-end processing complex. Emerging evidence suggests that Star-PAP may not require all canonical cleavage factors, while requiring additional proteins not present in the canonical 3′-end processing machinery. In the canonical mechanism, CPSF 160 binds the PAS, cooperates with CstF and CF Im. CPSF 160 then recruits PAPα through direct interaction at the cleavage site [26,28,29,45]. PAPα has low affinity for RNA substrate and lacks RNA binding specificity [100,114]; and no precise role of PAPα in the cleavage reaction has been defined. In contrast, Star-PAP directly binds pre-mRNA and plays a structural role to assemble the cleavage complex. Star-PAP directly interacts with CPSF 160 and 73 [19,24]. Star-PAP binding to the pre-mRNA and CPSF 160 recruits CPSF 160 to the PAS. CPSF 73 is then recruited to the cleavage site by its interaction with CPSF 160 and Star-PAP [24]. The mechanism of canonical PAP and Star-PAP mediated 3′-end processing is shown in Fig. 3.
Fig. 3.
A comparative model of the assembly of canonical and Star-PAP mediated 3′-end processing complexes. In the canonical model, CPSF 160 recognises the PAS and cooperates with CstF and CF Im and other factors to assemble a stable cleavage complex. The CPSF complex then recruits PAPα to the cleavage site by virtue of CPSF 160 direct interaction to PAPα (for a review see Refs. [1,4,5,7,8]). hFIP 1 also interacts with PAP to help position PAPα to the cleavage site. In case of Star-PAP mediated 3′-end processing, Star-PAP directly binds the target pre-mRNA UTR, a GC-rich sequence upstream of poly(A) signal, and helps recruit CPSF 160 and 73 subunits to the cleavage site to assemble a stable cleavage complex (see Refs. [20–22]). This complex subsequently excludes PAPα from the Star-PAP target UTRs.
Star-PAP has two RNA binding motifs: a Zinc Finger (ZF) and an RNA recognition motif (RRM), both required for mRNA binding [24]. This implies a complex binding motif of Star-PAP on its target RNA with multiple nucleotide elements that in combination interact with ZF, RRM, or both. This is consistent with the large Star-PAP footprint observed on targets, HO-1 and BIK UTR RNA [19,24]. The multiple binding elements could in turn enhance both specificity and flexibility for targeting specific pre-mRNAs. Moreover, Star-PAP and CPSF subunits 160 and 73 reconstitute cleavage of HO-1 UTR RNA in vitro. The resulting 3′-cleavage is specific but weak, suggesting that optimum in vivo cleavage requires other processing factors [24]. In contrast, no combinations of recombinant cleavage factors from the canonical mechanism could reconstitute in vitro cleavage reaction. This demonstrates that Star-PAP mediated 3′-end processing requires different sets of cleavage factors. In addition, due to low U/GU DSE on Star-PAP target mRNAs CstF which otherwise cooperates with CPSF-RNA binding in the canonical mechanism is likely dispensable in the Star-PAP mediated 3′-end processing mechanism [19]. Alternatively, different combinations of cleavage factors might function with specific PAPs (Star-PAP and PAPα) to regulate distinct target messages.
6. Implications of PAP diversity – Star-PAP vs canonical PAP
While the diversity of cellular PAPs is well known, the significance of PAP multiplicity is not clear. Multiple lines of evidence suggest the involvement of different PAPs in distinct cellular functions as illustrated by the functional specificities of various canonical isoforms and non-canonical PAPs. For example, ncPAPs such as hGLD2 or PAPD1 have specific roles in regulating cytoplasmic or mitochondrial mRNAs [118,121,126,135] while canonical PAPs such as PAPβ and PAPγ specifically regulate mRNAs during spermatogenesis and tumourigenesis respectively [12,15,17]. Moreover, longer canonical PAPα isoforms, I and IV exhibit tissue dependent expression [18]. On the other hand, Star-PAP regulates specific mRNA targets in the nucleus, which are otherwise inaccessible to PAPα [20,24]. Therefore, the regulation of distinct PAPs might differentially control expression of specific mRNAs.
6.1. Multiple types of 3′-end tail formation
There are two basic types of 3′-end tails reported so far: poly(A) tail and short terminal (U)-tail, both of which can have either destabilising or stabilising functions on RNA target. For example, poly(A) tails on mRNAs primarily confer stability to the transcript, while polyadenylation of aberrant tRNAs targets them for degradation [139,140]. Such destabilising function of poly(A) tail is well established in prokaryotic polyadenylation [186,187]. In general, polyadenosine tail present on mRNA 3′-ends can be either long poly(A) tail with stabilising function as in regular mRNA formation, or shorter (A) tail that destabilises RNA for degradation [9]. Further, small RNAs such as miRNAs as well as histone mRNAs are 3′-uridylated, apparently destabilising the RNA [123,124,188,189]. In contrast, oligo (U)-tailing of human U6 snRNA has a stabilising function [23,188]. In yeast, an additional type of 3′-end tail has been reported where the poly(A) tail is followed by a short U tract that results in degradation of the mRNA [158]. PAPα forms poly(A) tail at the 3′-end, and Star-PAP makes both poly(A) tails (like canonical PAPα) and also short terminal U tails (like a non-canonical PAP) to distinct target RNAs. Given the enzymatic character of Star-PAP and the number of target mRNAs it controls [20,23], there could be yet another type of a heterogeneous 3′-end tail having both As and Us in the cell. Such tails would have entirely different properties from that of either poly(A) or U tails [157,190].
6.2. Specificity for target mRNAs
It is clear that the two nuclear PAPs PAPα and Star-PAP control distinct mRNA targets in the nucleus. However, it is unclear why Star-PAP target mRNAs are specific to Star-PAP and not accessible to PAPα, and vice versa. A possibility for the specificity is the Star-PAP binding to its target pre-mRNA. Star-PAP footprint on its targets HO-1 and BIK pre-mRNA indicated a GC-rich sequence ~40–60 nucleotides upstream of the cleavage site [19,24] which is present on all Star-PAP target mRNAs [19]. RNA-compete analysis has also identified a specific Star-PAP (TUT1) binding oligonucleotide (-AUA-) motif [191]. This motif is present within the GC-rich sequence of Star-PAP footprints of all target mRNAs so far studied [19,24], suggesting that this motif might provide selectivity to Star-PAP for its target messages. However, this fails to explain why PAPα cannot process Star-PAP target messages even though intact canonical AAUAAA signal and cleavage site are present. Sequence analysis has shown a U/GU deplete DSE in Star-PAP target pre-mRNAs, which is critical for CstF binding [19]. Therefore, another possible explanation for PAPα exclusion from the Star-PAP regulated transcripts could be the low U/GU sequence (suboptimal DSE), which renders the pre-mRNA inaccessible to CstF, thus preventing the recruitment of PAPα. However, some of the Star-PAP target pre-mRNAs still possess DSE albeit weaker than the canonical counterparts [24]. Therefore, yet another explanation is the involvement of trans-acting factors that bind the pre-mRNA along with Star-PAP and confer specificity to Star-PAP targets. Like Star-PAP, other PAPs are also likely to have distinct sequence elements at the 3′-UTRs that determine the specificity of each PAP.
6.3. Signalling mediated regulation of polyadenylation
Star-PAP is regulated by signalling pathways through distinct kinases. Like Star-PAP, PAPα too is regulated by phosphorylation; however, the extracellular signal that triggers PAPα phopshoryation is unclear. Several serine and threonine residues within the extended C-terminus of PAPα are phosphorylated by cdc2-cyclin B [105,106]. During the mitotic phase of the cell cycle, this region is hyperphosphorylated to inhibit PAP activity, which is then reversed in the G1 phase [106]. Thus, this exemplifies a conditional or temporal regulation of PAP by phosphorylation as per cellular requirements. In addition, reports suggest that phosphorylation of PAPα by ERK kinase promotes the PAP activity [192]. PAP phosphorylation could also modulate the interaction with other cleavage factors to regulate distinct cellular functions. Thus, regulation of PAP could be an important mechanism for the cellular control of gene expression.
On the other hand, Star-PAP activity and its target gene expressions are stimulated by oxidative stress and nuclear PI4,5P2 [20,24]. The kinases and upstream signalling events modulating Star-PAP upon stress induction or DNA damage are well defined. Star-PAP associates with Ser/Thr kinase casein kinase I (CKI) [193,194], and protein kinase Cδ (PKCδ) [195–197]. Star-PAP is phosphorylated at the proline rich region (PRR) in the catalytic domain by CKI isoforms α and ε, which is critical for Star-PAP activity [152,153]. Knockdown of casein kinase or dephosphorylation of Star-PAP resulted in diminished Star-PAP activity. The PRR contains several putative CKI phosphorylation sites, and is likely to be phosphorylated at multiple sites. Both CKI isoforms together regulate 3′-processing of Star-PAP target HO-1 mRNA [153]. Oxidative stress treatment resulted in the induction of Star-PAP phosphorylation indicating the involvement of a stress signalling pathway coupled with phosphorylation to regulate Star-PAP.
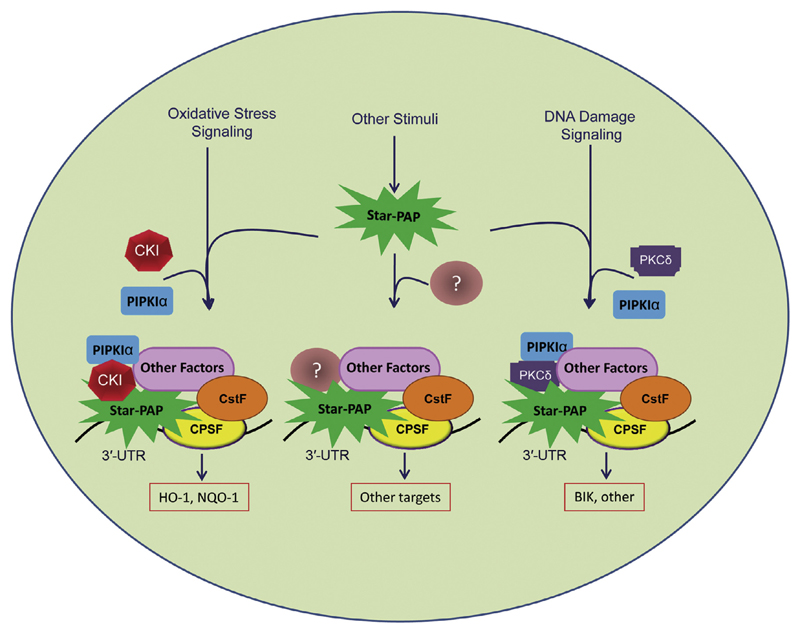
Unlike the CKI isoforms, PKCδ regulates Star-PAP activity downstream of DNA damage signalling [19]. PKCδ interacts with and phosphorylates Star-PAP. Interestingly, PKCδ is required for the DNA damage signal induced Star-PAP activity, but not for the oxidative stress induced pathway. While CKI and the stress induced pathway regulate genes involved in oxidative stress response such as HO-1 and NQO-1, DNA damage and PKCδ regulate the pro-apoptotic gene BIK. This signifies a signal mediated differential regulation of Star-PAP target genes. Thus, modulation of PAP by different signalling pathways regulates genes involved in distinct cellular functions suggesting that PAPs act as regulatory molecules that alter gene expression to mediate selective or conditional gene expression. Differential regulation of Star-PAP by distinct signalling pathways is depicted in Fig. 4.
Fig. 4.
Model for signal mediated regulation of Star-PAP to differentially control the 3′-end processing of distinct target messages. Various signal transduction components integrate into the Star-PAP 3′-end processing complex downstream of the signalling pathway, to regulate different target genes. In this model, casein kinase (isoforms α and ε) works downstream of oxidative stress to specifically regulate stress response genes HO-1 and NQO-1 through Star-PAP. On the other hand, PKCδ works in concert with DNA damage signal to regulate proapoptotic gene BIK through Star-PAP. The PKCδ mediated pathway is independent of oxidative stress regulation and vice versa. Thus, Star-PAP acts as a central regulatory molecule at the 3′-end of a gene that differentially controls the expression of target genes through various kinases and signalling molecules.
6.4. Alternative polyadenylation
The requirements of distinct cis-elements at the 3′-UTR of Star-PAP target genes have been discussed above. All nuclear pre-mRNA UTRs can have either Star-PAP specific or canonical PAP specific cis-element (poly(A) site). Reports indicate that more than 50% of mRNAs have multiple poly(A) sites at their 3′-UTRs [76]. It is likely that at least few of such mRNAs contain both Star-PAP and PAPα specific cis-elements at the 3′-UTR. This would result in alternate selection of poly(A) sites by poly(A) polymerases. The regulation of specific mRNAs or target 3′-UTRs by distinct canonical PAP isoforms has been reported in plants [117]. Several Star-PAP target genes identified in the microarray analysis also harbour more than one poly(A) site [20]. For example, NQO-1 has three poly(A) sites of which mRNA encoded by the most distal site is induced by the toxin dioxin.[198]. Knockdown of Star-PAP resulted in the loss of NQO-1 expression [20] suggesting that Star-PAP controls one or more poly(A) sites of NQO-1. In addition, expressions of some Star-PAP target genes are only partially diminished upon Star-PAP knockdown [20]. Such genes could represent a set of APA regulated genes where loss of Star-PAP regulated mRNA isoform is compensated by the expression of alternate isoforms from PAPα controlled poly(A) site(s) – a novel mechanism of APA site selection by PAPs.
7. PAP switch/PAP selection enigma at the 3′-end
It is evident that Star-PAP and PAPα assemble distinct 3′-end processing complexes. PAPα is not detected in the Star-PAP 3′-end processing complex and vice versa [20,24]. Intriguingly, there has been growing evidence that 3′-end processing factors are closely linked to the promoter, and at least in some cases may ride with RNA Pol II complex to 3′-end [199–204] (for review [205]). Evidence suggests that 3′-processing factor(s) such as CPSF are delivered to the promoter by transcription factor TFIID and then transferred to the elongating RNA pol II CTD [200,201]. Other studies also suggest the recruitment of 3′-end processing factors such as CPSF and CstF at the 5′-end [206]. Consistently, in yeast cleavage factors CFI and PFI have been detected with RNA Pol II starting at the promoter [201]. Since 3′-end processing factors exist in a tight complex, it is likely that they may all be detected in association with RNA Pol II either at the promoter, or after RNA Pol II clears the promoter (for reviews [205,207,208]). Since PAPs directly interact with CPSF [24,26], it is possible that PAPs associate with RNAP-CPSF complex during transcription. However, at this point there is no direct evidence that shows PAP association with RNA Pol II. Nevertheless, ChIP experiments have shown that yeast PAP1 is localised at the promoter, though it is preferentially bound at the 3′-UTR [209]. Thus, PAPs could join RNA Pol II via CPSF interaction during transcription elongation/initiation, or be recruited specifically to the 3′-end.
Both scenarios raise an important question: what determines the choice between the two PAPs (Star-PAP and canonical PAPα) to function at a particular 3′-UTR? There are two possible models for the PAP specificity at the 3′-UTR: PAP selection, or 3′-PAP switch. In the PAP selection model specific cis-elements on the 3′-UTR RNA select the required PAP. Alternatively, in the PAP switch model the canonical PAPα predominantly associates with 3′-RNA processing-transcription complex at the 3′-UTR of all pre-mRNAs. However, a PAP switch occurs at the 3′-UTR of Star-PAP target mRNAs, to Star-PAP due to its specific binding to mRNA to assemble a stable 3′-processing complex. This model is shown in Fig. 5. This mechanism of PAP selection/switch can be extrapolated to other PAP isoforms as well. However, at present what factors drive the PAP selection/switch remains to be determined.
Fig. 5.
3′-PAP switch/PAP selection model. In this model, a PAP switch/selection at the 3′-end of a gene favouring a particular PAP is determined by the cis-elements present on the target UTR RNA.
8. Conclusion
In recent years, much progress has been made towards the understanding of 3′-end processing mechanisms mediated by the two nuclear PAPs. Nevertheless, some key questions remain unaddressed, including the physiological significance for such variations in polyadenylation mechanisms. Indeed, the Star-PAP mediated 3′-processing complex could offer an advantage of selective regulation while employing canonical 3′-processing factors [24]. Star-PAP target genes such as HO-1 and NQO-1 are inducible stress response genes stimulated during oxidative stress [210–212]. The particular mechanism of Star-PAP dependent cleavage and its selective stimulation by oxidative stress will help home in and induce stress response genes while excluding global processing events that should remain unaffected during oxidative stress. Thus, temporal and/or specific stimulation of genes can occur through modulation of distinct PAPs, under different signalling conditions.
Current data demonstrate that diverse PAP populations function in the cell. Is the multiplicity of PAPs a cellular necessity or another functional redundancy in the cell? The answer is unknown but growing evidence suggests that the modulation of different PAPs regulate distinct cellular functions. For example, Star-PAP and PAPα are modulated by different kinases and signalling pathways to control expression of distinct target messages. Not only different PAPs but also distinct isoforms of canonical PAPα are differentially expressed, and are likely to regulate genes conditionally, or in a tissue dependent manner [12,17,18]. In conclusion, the existence of differentially regulated diverse PAP populations immensely benefits the cellular machinery for gene regulation. Although research over the last decade has considerably improved our knowledge on PAP functions, further studies are required to unravel how distinct PAPs attain specificity for their target transcripts and/or 3′-UTR selection.
Acknowledgements
We thank Fiona Ukken, C. Dustin Rubinstein (University of Wisconsin-Madison) and RSL Lab members for carefully reading the manuscript. We acknowledge the funding from Wellcome Trust-India Alliance (IA/I/1/505008). RSL is also an IYBA Fellow of the Department of Biotechnology, Government of India.
References
- [1].Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- [2].Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- [3].Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- [4].Proudfoot N, O’Sullivan J. Polyadenylation: a tail of two complexes. Curr Biol. 2002;12:R855–R857. doi: 10.1016/s0960-9822(02)01353-2. [DOI] [PubMed] [Google Scholar]
- [5].Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- [7].Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- [9].Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip. Rev RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- [10].Edmonds M. A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- [11].Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kashiwabara S, Zhuang T, Yamagata K, Noguchi J, Fukamizu A, Baba T. Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev Biol. 2000;228:106–115. doi: 10.1006/dbio.2000.9894. [DOI] [PubMed] [Google Scholar]
- [13].Kyriakopoulou CB, Nordvarg H, Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J Biol Chem. 2001;276:33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- [14].Le YJ, Kim H, Chung JH, Lee Y. Testis-specific expression of an intronless gene encoding a human poly(A) polymerase. Mol Cells. 2001;11:379–385. [PubMed] [Google Scholar]
- [15].Lee YJ, Lee Y, Chung JH. An intronless gene encoding a poly(A) polymerase is specifically expressed in testis. FEBS Lett. 2000;487:287–292. doi: 10.1016/s0014-5793(00)02367-x. [DOI] [PubMed] [Google Scholar]
- [16].Raabe T, Bollum FJ, Manley JL. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- [17].Topalian SL, Kaneko S, Gonzales MI, Bond GL, Ward Y, Manley JL. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol Cell Biol. 2001;21:5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhao W, Manley JL. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol. 1996;16:2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li W, Laishram RS, Ji Z, Barlow CA, Tian B, Anderson RA. Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIalpha and PKCdelta signaling. Mol Cell. 2012;45:25–37. doi: 10.1016/j.molcel.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- [21].Schmidt MJ, Norbury CJ. Polyadenylation and beyond: emerging roles for non-canonical poly(A) polymerases. Wiley Interdiscip Rev RNA. 2010;1:142–151. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- [22].Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- [23].Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Laishram RS, Anderson RA. The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. EMBO J. 2010;29:4132–4145. doi: 10.1038/emboj.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- [27].Bardwell VJ, Wickens M, Bienroth S, Keller W, Sproat BS, Lamond AI. Site-directed ribose methylation identifies 2′-OH groups in polyadenylation substrates critical for AAUAAA recognition and poly(A) addition. Cell. 1991;65:125–133. doi: 10.1016/0092-8674(91)90414-t. [DOI] [PubMed] [Google Scholar]
- [28].Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gilmartin GM, Nevins JR. Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol Cell Biol. 1991;11:2432–2438. doi: 10.1128/mcb.11.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ruegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- [32].Weiss EA, Gilmartin GM, Nevins JR. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991;10:215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lutz CS, Murthy KG, Schek N, O’Connor JP, Manley JL, Alwine JC. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- [34].Gilmartin GM, Fleming ES, Oetjen J. Activation of HIV-1 pre-mRNA 3′ processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gilmartin GM, Fleming ES, Oetjen J, Graveley BR. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- [36].Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- [39].Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jenny A, Hauri HP, Keller W. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol Cell Biol. 1994;14:8183–8190. doi: 10.1128/mcb.14.12.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J Biol Chem. 1997;272:26769–26779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- [42].Takagaki Y, MacDonald CC, Shenk T, Manley JL. The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc Natl Acad Sci USA. 1992;89:1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- [44].Takagaki Y, Manley JL. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- [45].Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- [46].Yang Q, Coseno M, Gilmartin GM, Doublie S. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 2011;19:368–377. doi: 10.1016/j.str.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yang Q, Gilmartin GM, Doublie S. The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5′ capping and splicing. RNA Biol. 2011;8(5):748–753. doi: 10.4161/rna.8.5.16040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Venkataraman K, Brown KM, Gilmartin GM. Analysis of a non-canonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- [51].He X, Khan AU, Cheng H, Pappas DL, Jr, Hampsey M, Moore CL. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem. 2009;284:22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kuhn U, Nemeth A, Meyer S, Wahle E. The RNA binding domains of the nuclear poly(A)-binding protein. J Biol Chem. 2003;278:16916–16925. doi: 10.1074/jbc.M209886200. [DOI] [PubMed] [Google Scholar]
- [56].Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim Biophys Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- [57].Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- [58].Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- [59].Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein–protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- [60].Kerwitz Y, Kuhn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, et al. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 2003;22:3705–3714. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Edmonds M, Abrams R. Polynucleotide biosynthesis: formation of a sequence of adenylate units from adenosine triphosphate by an enzyme from thymus nuclei. J Biol Chem. 1960;235:1142–1149. [PubMed] [Google Scholar]
- [62].Darnell JE, Wall R, Tushinski RJ. An adenylic acid-rich sequence in messenger RNA of HeLa cells and its possible relationship to reiterated sites in DNA. Proc Natl Acad Sci USA. 1971;68:1321–1325. doi: 10.1073/pnas.68.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Edmonds M, Vaughan MH, Jr, Nakazato H. Polyadenylic acid sequences in the heterogeneous nuclear RNA and rapidly-labeled polyribosomal RNA of HeLa cells: possible evidence for a precursor relationship. Proc Natl Acad Sci USA. 1971;68:1336–1340. doi: 10.1073/pnas.68.6.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Winters MA, Edmonds M. A poly(A) polymerase from calf thymus. Purification and properities of the enzyme. J Biol Chem. 1973;248:4756–4762. [PubMed] [Google Scholar]
- [65].Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Keller RW, Kuhn U, Aragon M, Bornikova L, Wahle E, Bear DG. The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. J Mol Biol. 2000;297:569–583. doi: 10.1006/jmbi.2000.3572. [DOI] [PubMed] [Google Scholar]
- [68].Palaniswamy V, Moraes KC, Wilusz CJ, Wilusz J. Nucleophosmin is selectively deposited on mRNA during polyadenylation. Nat Struct Mol Biol. 2006;13:429–435. doi: 10.1038/nsmb1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sagawa F, Ibrahim H, Morrison AL, Wilusz CJ, Wilusz J. Nucleophosmin deposition during mRNA 3′ end processing influences poly(A) tail length. EMBO J. 2011;30:3994–4005. doi: 10.1038/emboj.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508(7494):66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wickens M, Belasco JG, Jacobson A. Changes in the length of poly (A) tails and their effects on mRNA translation and turnover. Post-transcriptional Control of Gene, NATO ASI Series. 1996;97:45–55. [Google Scholar]
- [72].Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- [73].Danckwardt S, Hentze MW, Kulozik AE. 3’ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- [76].Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ihara H, Tsutsuki H, Ida T, Kozaki S, Tsuyama S, Moss J. Alternative polyadenylation sites of human endothelial nitric oxide synthase mRNA. Biochem Biophys Res Commun. 2007;363:146–152. doi: 10.1016/j.bbrc.2007.08.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Misquitta CM, Iyer VR, Werstiuk ES, Grover AK. The role of 3′-untranslated region (3′-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol Cell Biochem. 2001;224:53–67. doi: 10.1023/a:1011982932645. [DOI] [PubMed] [Google Scholar]
- [80].Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, et al. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10:327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- [84].Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lou H, Neugebauer KM, Gagel RF, Berget SM. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- [88].Morris AR, Bos A, Diosdado B, Rooijers K, Elkon R, Bolijn AS, et al. Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res. 2012;18:5256–5266. doi: 10.1158/1078-0432.CCR-12-0543. [DOI] [PubMed] [Google Scholar]
- [89].Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27(21):2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, et al. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980;20:313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- [92].Rogers J, Early P, Carter C, Calame K, Bond M, Hood L, et al. Two mRNAs with different 3′ ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980;20:303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- [93].Gunderson SI, Vagner S, Polycarpou-Schwarz M, Mattaj IW. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- [94].Ballantyne S, Bilger A, Astrom J, Virtanen A, Wickens M. Poly (A) polymerases in the nucleus and cytoplasm of frog oocytes: dynamic changes during oocyte maturation and early development. RNA. 1995;1:64–78. [PMC free article] [PubMed] [Google Scholar]
- [95].Ryner LC, Takagaki Y, Manley JL. Multiple forms of poly(A) polymerases purified from HeLa cells function in specific mRNA 3′-end formation. Mol Cell Biol. 1989;9:4229–4238. doi: 10.1128/mcb.9.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wahle E, Martin G, Schiltz E, Keller W. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J. 1991;10:4251–4257. doi: 10.1002/j.1460-2075.1991.tb05003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Aravind L, Koonin EV. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Holm L, Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- [99].Martin G, Jeno P, Keller W. Mapping of ATP binding regions in poly(A) polymerases by photoaffinity labeling and by mutational analysis identifies a domain conserved in many nucleotidyltransferases. Protein Sci. 1999;8:2380–2391. doi: 10.1110/ps.8.11.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- [101].Martin G, Keller W, Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Raabe T, Murthy KG, Manley JL. Poly(A) polymerase contains multiple functional domains. Mol Cell Biol. 1994;14:2946–2957. doi: 10.1128/mcb.14.5.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Davies JF, 2nd, Almassy RJ, Hostomska Z, Ferre RA, Hostomsky Z. 2.3 A crystal structure of the catalytic domain of DNA polymerase beta. Cell. 1994;76:1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- [104].Bard J, Zhelkovsky AM, Helmling S, Earnest TN, Moore CL, Bohm A. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science. 2000;289:1346–1349. doi: 10.1126/science.289.5483.1346. [DOI] [PubMed] [Google Scholar]
- [105].Bond GL, Prives C, Manley JL. Poly(A) polymerase phosphorylation is dependent on novel interactions with cyclins. Mol Cell Biol. 2000;20:5310–5320. doi: 10.1128/mcb.20.14.5310-5320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhao W, Manley JL. Deregulation of poly(A) polymerase interferes with cell growth. Mol Cell Biol. 1998;18:5010–5020. doi: 10.1128/mcb.18.9.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- [108].Colgan DF, Murthy KG, Zhao W, Prives C, Manley JL. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- [110].Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- [111].Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- [112].Perumal K, Sinha K, Henning D, Reddy R. Purification, characterization, and cloning of the cDNA of human signal recognition particle RNA 3′-adenylating enzyme. J Biol Chem. 2001;276:21791–21796. doi: 10.1074/jbc.M101905200. [DOI] [PubMed] [Google Scholar]
- [113].Tupler R, Perini G, Green MR. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- [114].Wahle E. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1991;266:3131–3139. [PubMed] [Google Scholar]
- [115].Gebauer F, Richter JD. Cloning and characterization of a Xenopus poly(A) polymerase. Mol Cell Biol. 1995;15:3460. doi: 10.1128/mcb.15.3.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Nordvarg H. Functional significance of multiple poly(A) polymerases (PAPs) Acta Univ Ups. 2002;1140:1–51. [Google Scholar]
- [117].Vi SL, Trost G, Lange P, Czesnick H, Rao N, Lieber D, et al. Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc Natl Acad Sci USA. 2013;110:13994–13999. doi: 10.1073/pnas.1303967110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- [120].Ojala D, Montoya J, Attardi G. TRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- [121].Nagaike T, Suzuki T, Ueda T. Polyadenylation in mammalian mitochondria: insights from recent studies. Biochim Biophys Acta. 2008;1779:266–269. doi: 10.1016/j.bbagrm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- [122].Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- [123].Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Schmidt MJ, West S, Norbury CJ. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011;17:39–44. doi: 10.1261/rna.2252511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008;135:1969–1979. doi: 10.1242/dev.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci USA. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Glahder JA, Norrild B. Involvement of hGLD-2 in cytoplasmic polyadenylation of human p53 mRNA. APMIS. 2011;119:769–775. doi: 10.1111/j.1600-0463.2011.02804.x. [DOI] [PubMed] [Google Scholar]
- [129].Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- [130].Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze M. Translational control in developmental decisions. In: Sonenberg N, et al., editors. Translational Control of Gene Expression. Cold Spring Harbor Lab. Press; Plainview, NY: 2000. pp. 295–370. [Google Scholar]
- [131].Villalba A, Coll O, Gebauer F. Cytoplasmic polyadenylation and translational control. Curr Opin Genet Dev. 2011;21:452–457. doi: 10.1016/j.gde.2011.04.006. [DOI] [PubMed] [Google Scholar]
- [132].Burns DM, D’Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473(7345):105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- [134].Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Dickson KS, Thompson SR, Gray NK, Wickens M. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J Biol Chem. 2001;276:41810–41816. doi: 10.1074/jbc.M103030200. [DOI] [PubMed] [Google Scholar]
- [136].Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Rammelt C, Bilen B, Zavolan M, Keller W. PAPD5, a non-canonical poly(A) polymerase with an unusual RNA-binding motif. RNA. 2011;17:1737–1746. doi: 10.1261/rna.2787011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–111. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Berndt H, Harnisch C, Rammelt C, Stohr N, Zirkel A, Dohm JC, et al. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012;18(5):958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Blahna MT, Jones MR, Quinton LJ, Matsuura KY, Mizgerd JP. Terminal uridyltransferase enzyme Zcchc11 promotes cell proliferation independent of its uridyltransferase activity. J Biol Chem. 2011;286:42381–42389. doi: 10.1074/jbc.M111.259689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- [145].Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 2010;20:25–35. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Bunce MW, Bergendahl K, Anderson RA. Nuclear PI(4,5)P(2): a new place for an old signal. Biochim Biophys Acta. 2006;1761:560–569. doi: 10.1016/j.bbalip.2006.03.002. [DOI] [PubMed] [Google Scholar]
- [151].Doughman RL, Firestone AJ, Anderson RA. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J Membr Biol. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- [152].Gonzales ML, Mellman DL, Anderson RA. CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J Biol Chem. 2008;283:12665–12673. doi: 10.1074/jbc.M800656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Laishram RS, Barlow CA, Anderson RA. CKI isoforms alpha and epsilon regulate Star-PAP target messages by controlling Star-PAP poly(A) polymerase activity and phosphoinositide stimulation. Nucleic Acids Res. 2011;39:7961–7973. doi: 10.1093/nar/gkr549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Newman A. RNA splicing. Activity in the spliceosome. Curr Biol. 1994;4:462–464. doi: 10.1016/s0960-9822(00)00104-4. [DOI] [PubMed] [Google Scholar]
- [155].Trippe R, Richly H, Benecke BJ. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur J Biochem. 2003;270:971–980. doi: 10.1046/j.1432-1033.2003.03466.x. [DOI] [PubMed] [Google Scholar]
- [156].Trippe R, Sandrock B, Benecke BJ. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by non-canonical poly(A) polymerases. Mol Cell Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3′–5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013;32:1855–1868. doi: 10.1038/emboj.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Fitzgerald M, Shenk T. The sequence 5′-AAUAAA-3′ forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981;24:251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- [163].Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- [165].Orkin SH, Cheng TC, Antonarakis SE, Kazazian HH., Jr Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 1985;4:453–456. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Bennett CL, Brunkow ME, Ramsdell F, O’Briant KC, Zhu Q, Fuleihan RL, et al. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA->AAUGAA) leads to the IPEX syndrome. Immunogenetics. 2001;53:435–439. doi: 10.1007/s002510100358. [DOI] [PubMed] [Google Scholar]
- [167].Wahle E, Keller W. The biochemistry of polyadenylation. Trends Biochem Sci. 1996;21:247–250. [PubMed] [Google Scholar]
- [168].Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Chen F, MacDonald CC, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Danckwardt S, Gehring NH, Neu-Yilik G, Hundsdoerfer P, Pforsich M, Frede U, et al. The prothrombin 3’end formation signal reveals a unique architecture that is sensitive to thrombophilic gain-of-function mutations. Blood. 2004;104:428–435. doi: 10.1182/blood-2003-08-2894. [DOI] [PubMed] [Google Scholar]
- [172].Danckwardt S, Hartmann K, Gehring NH, Hentze MW, Kulozik AE. 3′ end processing of the prothrombin mRNA in thrombophilia. Acta Haematol. 2006;115:192–197. doi: 10.1159/000090934. [DOI] [PubMed] [Google Scholar]
- [173].Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, et al. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- [174].Chou ZF, Chen F, Wilusz J. Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res. 1994;22:2525–2531. doi: 10.1093/nar/22.13.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- [176].McLauchlan J, Gaffney D, Whitton JL, Clements JB. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 1985;13:1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Moreira A, Wollerton M, Monks J, Proudfoot NJ. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Prescott J, Falck-Pedersen E. Sequence elements upstream of the 3′ cleavage site confer substrate strength to the adenovirus L1 and L3 polyadenylation sites. Mol Cell Biol. 1994;14:4682–4693. doi: 10.1128/mcb.14.7.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H’ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Bagga PS, Ford LP, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Dalziel M, Nunes NM, Furger A. Two G-rich regulatory elements located adjacent to and 440 nucleotides downstream of the core poly(A) site of the intronless melanocortin receptor 1 gene are critical for efficient 3′ end processing. Mol Cell Biol. 2007;27:1568–1580. doi: 10.1128/MCB.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Cohen SN. Surprises at the 3′ end of prokaryotic RNA. Cell. 1995;80:829–832. doi: 10.1016/0092-8674(95)90284-8. [DOI] [PubMed] [Google Scholar]
- [187].Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu Rev Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- [188].Chen Y, Sinha K, Perumal K, Reddy R. Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA. 2000;6:1277–1288. doi: 10.1017/s1355838200000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- [190].Molloy GR. The isolation and characterization of HeLa cell messenger RNA-like molecules containing uridylic acid-rich oligonucleotide sequences. J Biol Chem. 1980;255:10375–10381. [PubMed] [Google Scholar]
- [191].Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Lee SH, Choi HS, Kim H, Lee Y. ERK is a novel regulatory kinase for poly(A) polymerase. Nucleic Acids Res. 2008;36:803–813. doi: 10.1093/nar/gkm1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- [194].Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- [195].Gschwendt M. Protein kinase C delta. Eur J Biochem. 1999;259:555–564. doi: 10.1046/j.1432-1327.1999.00120.x. [DOI] [PubMed] [Google Scholar]
- [196].Kikkawa U, Matsuzaki H, Yamamoto T. Protein kinase C delta (PKC delta): activation mechanisms and functions. J Biochem. 2002;132:831–839. doi: 10.1093/oxfordjournals.jbchem.a003294. [DOI] [PubMed] [Google Scholar]
- [197].Reyland ME. Protein kinase Cdelta and apoptosis. Biochem Soc Trans. 2007;35:1001–1004. doi: 10.1042/BST0351001. [DOI] [PubMed] [Google Scholar]
- [198].Jaiswal AK, McBride OW, Adesnik M, Nebert DW. Human dioxin-inducible cytosolic NAD(P)H:menadione oxidoreductase. cDNA sequence and localization of gene to chromosome 16. J Biol Chem. 1988;263:13572–13578. [PubMed] [Google Scholar]
- [199].Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- [200].Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- [201].Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, et al. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- [202].O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, et al. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- [203].Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- [206].Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108(501–1):2. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- [208].Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- [209].Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P. The spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol Cell Biol. 2012;32:1321–1331. doi: 10.1128/MCB.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Maines MD, Gibbs PE. 30 some years of heme oxygenase: from a molecular wreckleing ball to a mesmerizing trigger of cellular events. Biochem Biophys Res Commun. 2005;338:568–577. doi: 10.1016/j.bbrc.2005.08.121. [DOI] [PubMed] [Google Scholar]
- [211].Otterbein LE, Choi AM. Heme oxygenase: colors of defence against cellular stress. Am J Physiol Lung Cell Mol Physiol. 2000;279:1029–1037. doi: 10.1152/ajplung.2000.279.6.L1029. [DOI] [PubMed] [Google Scholar]
- [212].Ryter S, Otterbein LE, Morse D, Choi AM. Heme oxygenase/carbon monoxide signaling pathways: regulation and functional significance. Mol Cell Biochem. 2002;234:249–263. doi: 10.1023/A:1015957026924. [DOI] [PMC free article] [PubMed] [Google Scholar]