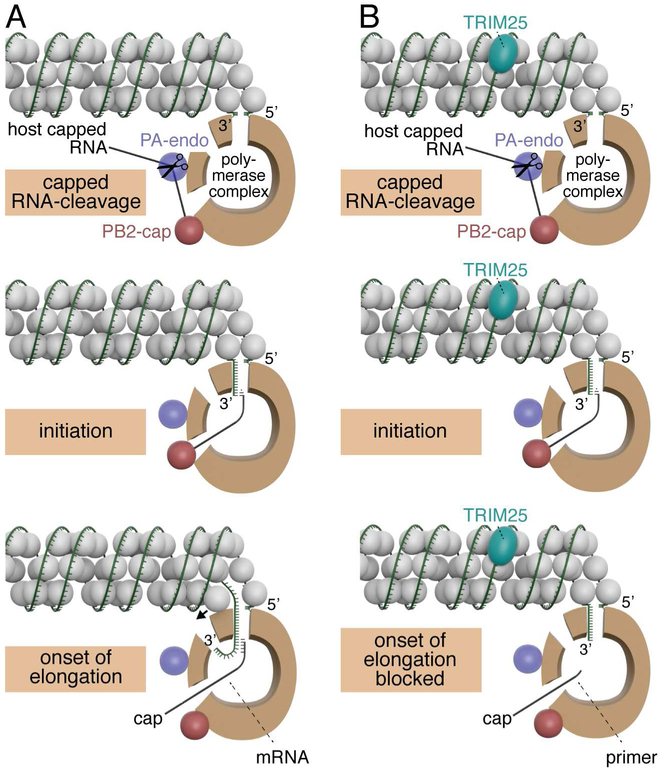
Figure 7. Model of the mechanism of TRIM25 inhibition.
Models for capped RNA-primed viral mRNA synthesis in the absence (panel A) and presence (panel B) of TRIM25. The relative sizes of NP (grey balls), TRIM25 (aqua) and the viral polymerase (brown) correspond to their relative molecular weights. The three protein subunits of the polymerase (PB1, PB2 and PA) make multiple interactions with each other, but these interactions are not shown in this model. Instead, the main body of the polymerase is represented as a brown “donut,” and entry and exit sites used during viral mRNA synthesis are depicted. The “donut hole” contains the catalytic site for nucleotide addition. The PB2 cap-binding site and PA endonuclease are located at one end of the main body of the polymerase, as depicted by the red and purple balls in this model. The capped RNA-cleavage and initiation steps are identical in the absence and presence of TRIM25. In contrast, the onset of RNA chain elongation is blocked in the presence of TRIM25, as discussed in the text. In the lower left panel, a small arrow indicates that NP has to be displaced in order for the RNA chain to enter the polymerase and be copied. These models contain aspects of the polymerase models shown in figure 3 of (Velthuis and Fodor, 2016).