Figure 2.
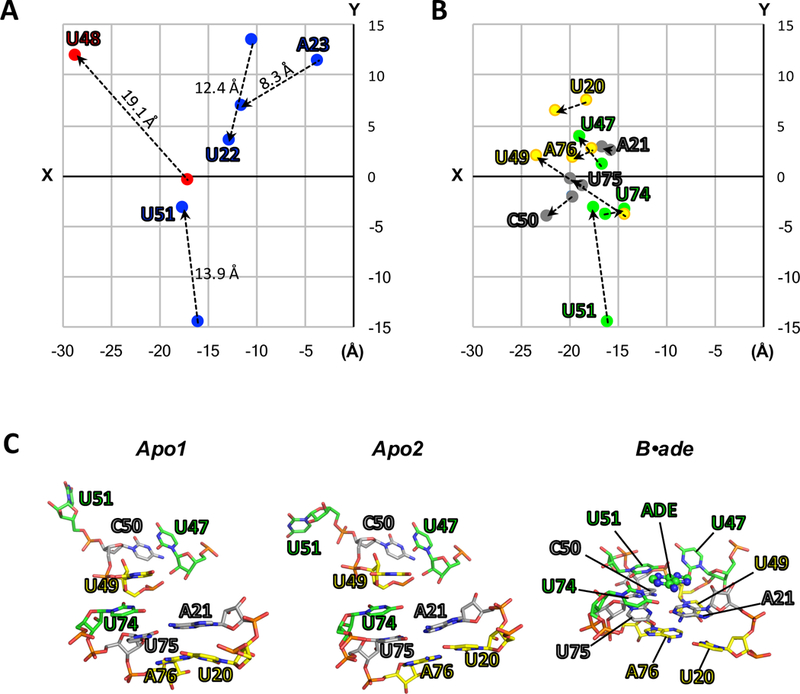
(A-B) Two-dimensional projections (XY-plane) of the structural coordinates (Å) of key residues for apo2 and B•ade. The structures of apo2 (PDB:5E54) and B•ade (PDB:4TZX) were aligned as in Fig. 1, and the XY-coordinates for O2 atoms (pyrimidines) or N6 atoms (purines) were taken directly from their respective PDB files and plotted. (A) “Swinging residues” that flip toward (blue) or away (red) from the ligand-binding pocket upon conversion to B•ade. The direction of movement is also indicated by dotted lines. Values reported above the dotted lines correspond to the distances in three dimensions. (B) Residues involved in the three ligand-facilitated base triples: U20-U49-A76 (yellow), A21-C50-U75 (grey), and U47-U51-U74-ade (green). (C) In the absence of ligand (apo1 and apo2), the base-triple interactions are broken. Ligand binding facilitates the formation of the base triples, as observed in the crystal structure of ligand-bound rA71 (PDB:4TZX). These interactions include ligand recognition by U74 and anchoring of P1 (residues U75 and A76) and the hinge (U20 and A21) via the latch (residues U47, U49, C50 and U51), ultimately leading to the stabilization of the P1 helix.
