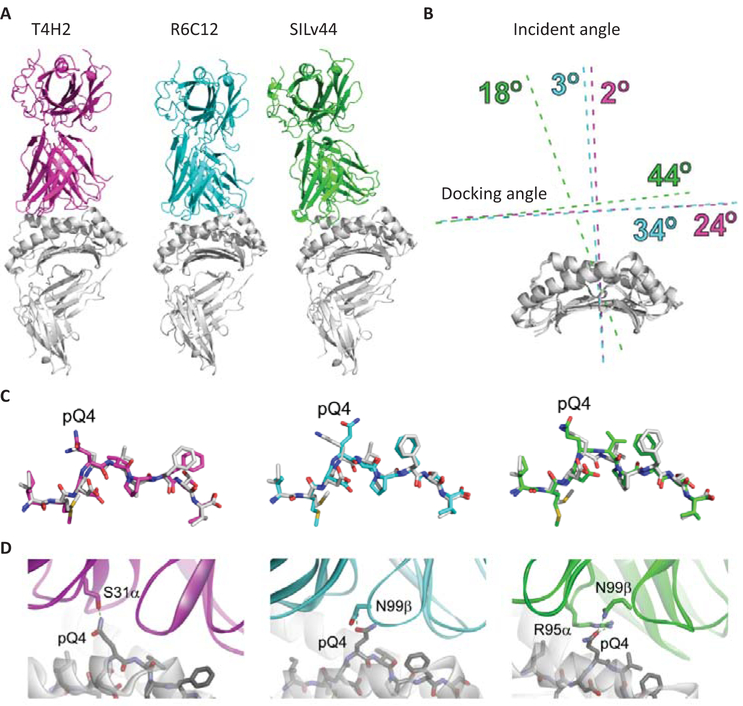
Fig. 5.
Structural models of the T4H2, R6C12, and SILv44 TCRs with gp100209–2M/HLA-A2. (A) Overview of the complexes. In the three models, the T4H2 complex is magenta, R6C12 is cyan, and SILv44 is green (this coloring is maintained for all panels). The orientation of the peptide/MHC is the same in all three images. The altered binding mode predicted for SILv44 is clearly apparent. (B) Quantification of TCR binding modes for the three complexes. The primary difference is in the incident angle, which is 18° off the vertical for SILv44, but 3° for R6C12 and 2° for T4H2. (C) Conformations of the modeled peptides in the three TCR-peptide/MHC models, compared to the conformation of the peptide in the unbound gp100209–2M/HLA-A2 crystal structure (white). The overall conformations are very similar, except for that with R6C12, in which pGln4 is predicted to be positioned more centrally in the TCR-peptide/MHC interface. (D) Engagement of pGln4 in the three TCR-peptide/MHC models.