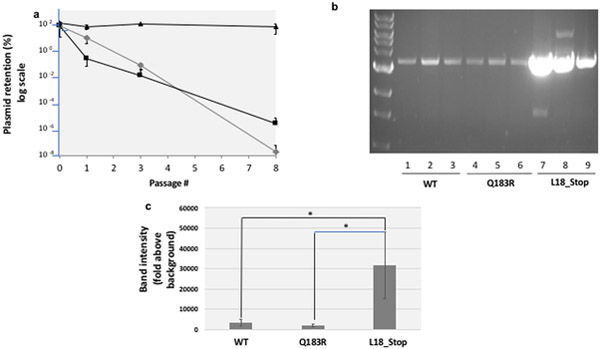
Fig. 2. The pGFPuv retention model of plasmid stability.
a. Plasmid stability. Cells transformed with plasmids harboring wild-type pGFPuv (grey diamonds), a Q183R mutant of GFP (which abolishes fluorescence, black squares), and a L18* truncated mutant (black triangles) were washed from plates with carbenicillin, normalized for cell number and expanded in liquid culture in the presence of carbenicillin. After the first passage (passage 0), these cells were passaged 8 more times through a 1:105 dilution and growth for 22h in LB at 37˚C in the absence of carbenicillin. To monitor plasmid retention, at specific time-points cells were withdrawn, serially diluted and plated in the presence or absence of carbenicillin. CFUs arising after 16 hours incubation at 37˚C were quantified. b Plasmid copy number. Plasmids from a normalized number of cells bearing the WT pGFPuv plasmid, the Q183R (non-fluorescent) mutant and the L18_Stop (truncated) mutant were recovered, linearized, and run on an agarose gel as described in the Methods. c Gel band density quantification. Band intensities in the gel shown in section b were measured by densitometry using Image J software, with fixed area size for all the samples, and averaged. Error bars represent the standard deviation for three replicates. The difference between L18_Stop and WT and between L18_Stop and Q183R was confirmed by a 2-tailed unpaired Student t test (p<0.05) (asterisks).