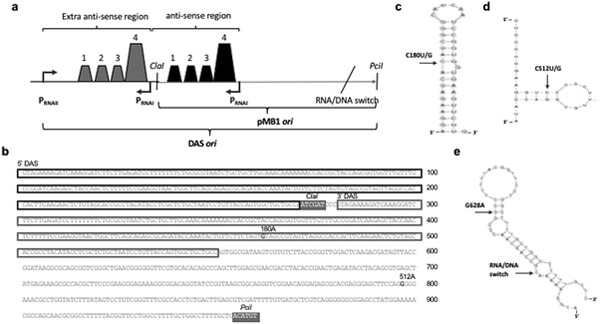
Fig. 6. Map of modifications increasing pMB1 stability.
a Duplicated antisense sequence Schematic showing the duplication of antisense sequence (DAS). Shown are promoters PRNAI and PRNAII; the stem-loop structures that are important for antisense regulation; the two restriction sites used for cloning of the plasmid ori mutants; and the position of the RNA/DNA switch. b. DAS ori sequence. Duplicated sequences are boxed; mutations tested for increased stability are highlighted in bold, denoting nucleotide positions (relative to the start of the ori following standard ColE1 numbering (Ohmori and Tomizawa, 1979)) and base pair substitutions. The two restriction sites used for cloning of the plasmid ori mutants are denoted and highlighted in grey boxes. c Location of the G180A/C mutation. This mutation is mapped on a secondary structure for stem-loop number four of RNAII, which comprises nucleotides 172 through 207. The secondary structure shown is the most stable one predicted using the mfold program (Zuker, 2003), with a ΔG=−12.20 kcal/mol. d Location of the G512A/C mutation. This mutation is mapped on a secondary structure for hairpin 2 of RNAII, which comprises nucleotides 508 through 525. The model shown is the most stable one predicted using the mfold program (Zuker, 2003), with a ΔG=−7.3 kcal/mol. e Location of the G628A mutation. This mutation is mapped on a secondary structure for single-stranded DNA in area surrounding the RNA/DNA switch. The model shown is the most stable one predicted using the mfold program (Zuker, 2003), with a ΔG=−11.27 kcal/mol.