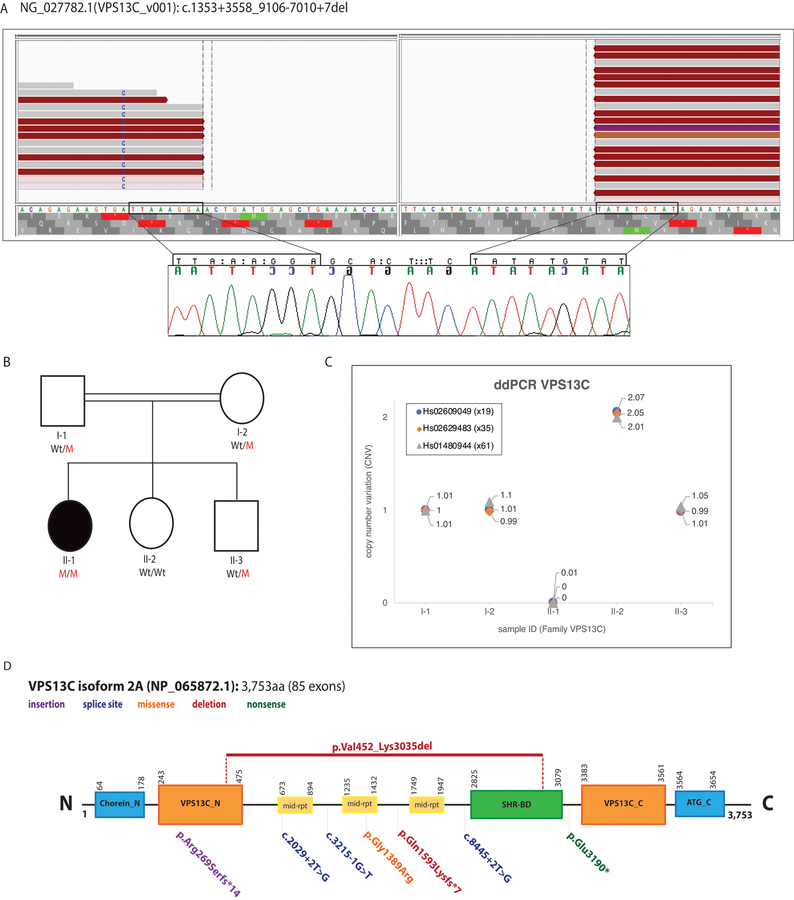
Fig 1.
(A) WGS reads (Top) and Sanger chromatograms (Bottom) illustrating the large homozygous VPS13C deletion (Blank area, no reads) identified in an isolated patient with parkinsonism (https://databases.lovd.nl/shared/variants/0000368891). The WGS reads were visualized using the IGV tool. The Sanger chromatograms show the evidence of a 7-bp insertion between the deletion breakpoints (nucleotides between rectangles). The breakpoints of the deletion are highlighted with rectangles. (B) Pedigree of the family with parkinsonism and sensorimotor polyneuropathy. Wt/M indicates heterozygous carrier for the VPS13C large deletion; M/M indicates homozygous carrier; and Wt/Wt indicates non-carrier. Affected patient is represented with a black circle. (C) CNV plots of VPS13C exons 19, 35, and 61 obtained through ddPCR (QX100 system, Bio-rad) and corresponding taqman assays (Applied Biosystems) are shown. Homozygous Wt alleles are represented with a CNV score of 2 or close to 2, meaning both alleles carry a copy of the exon under investigation; heterozygous mutant alleles are represented with a CNV score of 1 or close to 1, meaning only one allele carry a copy of the exon while the exon is absent in the other allele; homozygous mutant alleles are represented with a CNV score of 0, meaning the exon under investigation is absent in both alleles. Only subject II-1 is homozygous carrier for the identified VPS13C deletion. (D): VPS13C protein (NP_065872.1) structure located at chromosome 15q22.2 and predicted by SMART (http://smart.embl-heidelberg.de). Functional domains along with reported VPS13C mutations are shown. The VPS13C mutation identified in this study is shown at the top of the protein. Chorein_N: N terminal region of Chorein, VPS13C_N: N terminal domain, mid-rpt: VPS13_mid_repeats, SHR-BD: SHR binding domain, VPS13C_C: C terminal domain, ATG_C: Autophagy related protein C terminal domain.