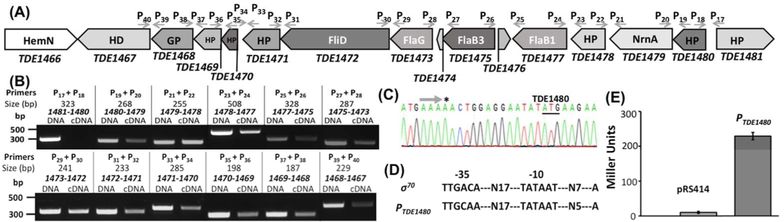
Figure 1. TDE1473 is located at a large gene cluster that is regulated by a σ70-like promoter.
(A) A diagram showing the genes adjacent to TDE1473. Arrows represent the relative positions and orientations of RT-PCR primers that span the intergenic regions between individual orfs. (B) RT-PCR analysis. For each pair of primers, chromosomal DNA was used as a positive control. The numbers below the primers are predicted sizes of RT-PCR and PCR products. (C) 5ˋ-RLM-RACE analysis. The arrow shows the sequencing direction and an asterisk (*) indicates the transcriptional start site. (D) Sequence comparison between the E. coli ſ70 promoter and the promoter sequence upstream of TDE1480 (designated as PTDE1480). (E) Transcriptional analysis of PTDE1480 using lacZ as a reporter in E. coli. For this assay, PTDE1480 was fused to the promoterless lacZ gene in the pRS414 plasmid. The promoterless pRS414 was used as a negative control. β-galactosidase activity was measured and expressed as the average Miller units of triplicate samples from two independent experiments, as previously described (Bian et al., 2013). All the primers used here are listed in Table 1.