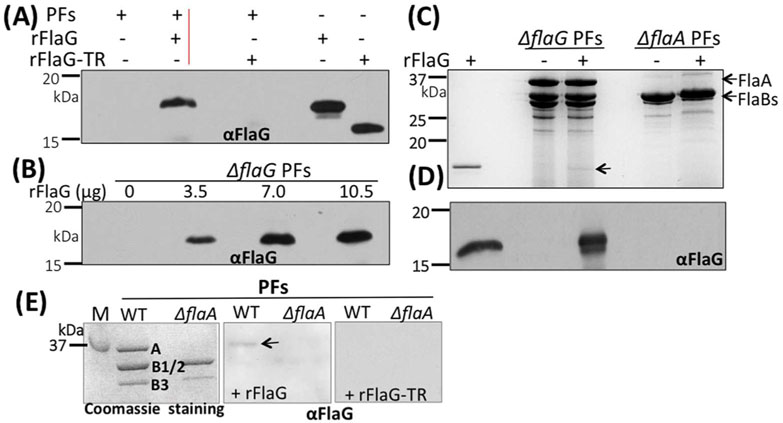
Figure 8. FlaG binds to the PFs in a FlaA-dependent manner.
(A)& (B) rFlaG binds to the filaments in a dose-dependent manner. For these experiments, different amounts of rFlaG as labeled were co-incubated with the PFs (18 μg) isolated from ΔflaG for 16 hrs, washed three times with PBS, and then subjected to SDS-PAGE, followed by immunoblotting with ⟨FlaG. (C) rFlaG fails to bind to the PFs isolated from the flaA mutant (ΔflaA). Similar to the above experiments, here rFlaG was co-incubated with the PFs isolated from ΔflaG and ΔflaA strains, and then subjected to SDS-PAGE, followed by Coomassie blue staining (top image) or immunoblotting with αFlaG (lower image). Arrow points to rFlaG detected. (D) Far-western blotting analysis. The PFs isolated from the wild type and ΔflaA strains were separated by SDS-PAGE (left image), transferred to PVDF membranes, incubated with rFlaG (middle image) or rFlaG-TR (C-terminus 15 aa were truncated, right image), washed 4 times with T-PBS, and finally probed with αFlaG. The blots were developed using a horseradish peroxidase secondary antibody with an enhanced ECL, as previously described (Kurniyati et al., 2017). Arrow points to FlaA detected. M: marker; A: FlaA; B1/2: FlaB1, FlaB2; B3: FlaB3.