Figure 6. Identification of L3mbtl1 Target Genes.
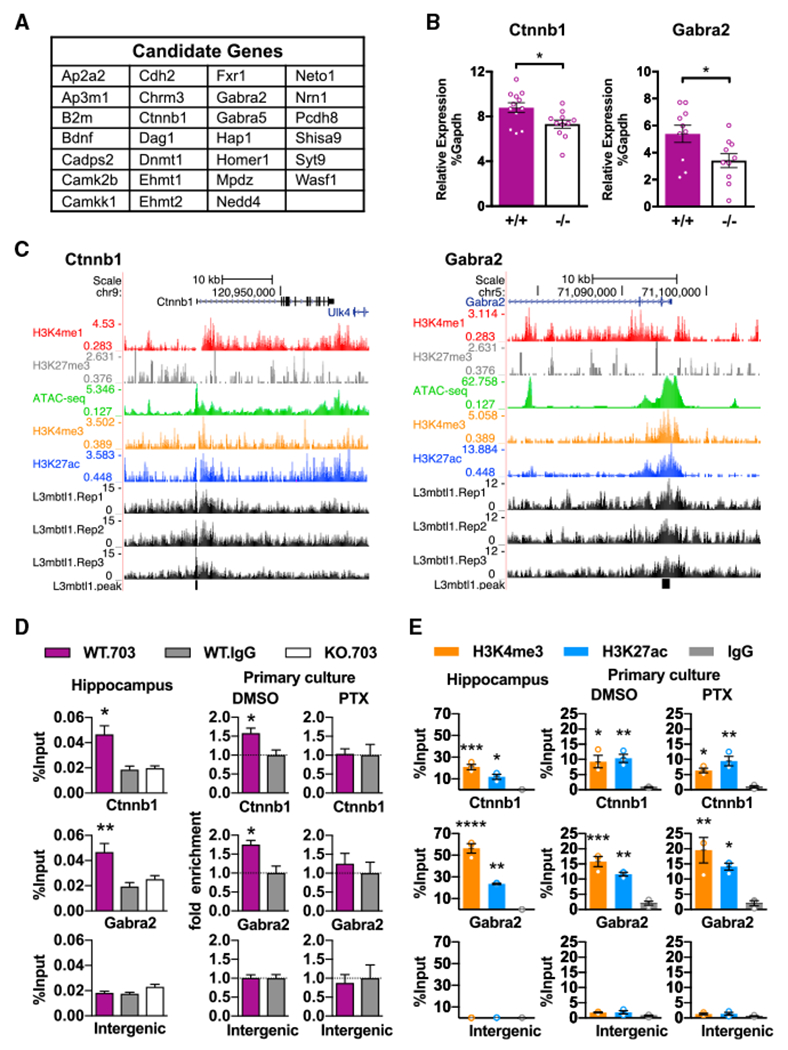
(A) List of selected putative L3mbtl1 target genes that have known functions in synaptic transmission and/or homeostatic synaptic plasticity. See also Tables S1 and S3 and Figure S6.
(B) Quantification of Ctnnb1 and Gabra2 RNA expression by qPCR. Ctnnb1 and Gabra2 expression relative to Gapdh were measured from hippocampal primary cultures prepared from wild-type (+/+) and L3mbtl1 KO (−/−) mice. **p < 0.01, *p<0.5; Student’s t test. N = 8–12 independent cultures.
(C) L3mbtl1-binding events at promoter regions of Ctnnb1 (left) and Gabra2 (right) genes obtained from ChIP-seq of P7 mouse hippocampus. ChlP-seq tracks for L3mbtl1 from wild-type samples (L3mbtl1.Rep1–3), various histone markers, and ATAC-seq tracks (GEO GSE63137) are shown. Black blocks indicate signal peaks for L3mbtl1-bound regions.
(D) L3mbtl1 binding validated by direct ChlP-qPCR assays at Ctnnb1 and Gabra2 promoter regions. ChIP was performed from P7 hippocampus (left) or primary cultures treated with DMSO (middle) or PTX (right) for 24 hr. Note that PTX treatment abolished L3mbtl1 binding in primary cultures. A one-way ANOVA test was used for the hippocampus and Student’s t test for primary culture. N = 3 biological replicates.
(E) Enrichment for H3K4me3 and H3K27ac validated by direct ChIP-qPCR assays at Ctnnb1 and Gabra2 promoter regions. ChIP was performed from P7 hippocampus (left) or primary cultures treated with DMSO (middle) or PTX (right). Note that PTX treatment did not change H3K4me3 or H3K27ac profiles at promoter regions. One-way ANOVA test. N = 3 biological replicates.
****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.5. Data shown are means ± SEM.
