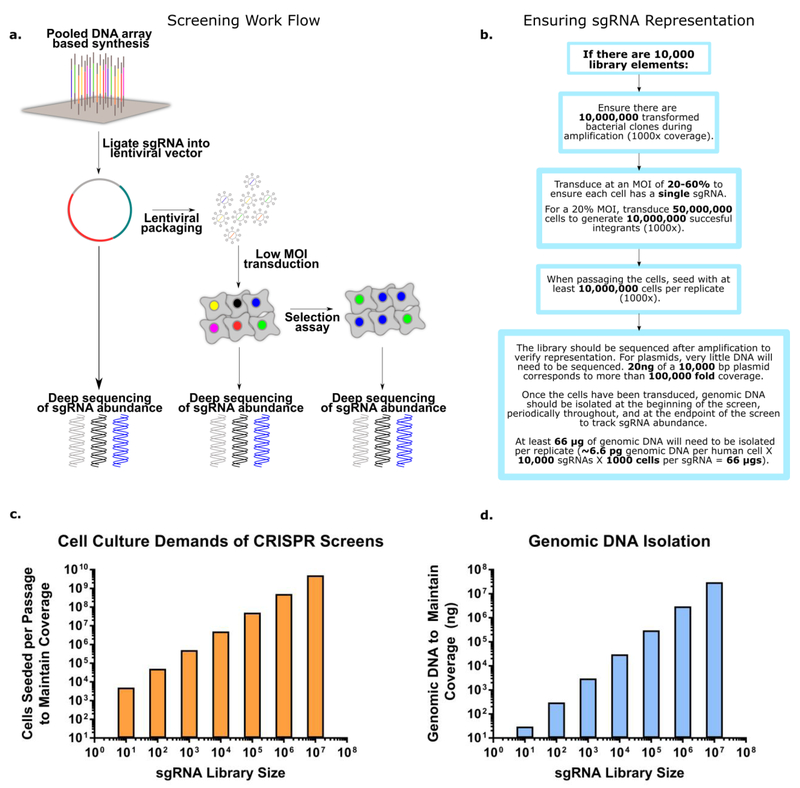
Figure 2. Mechanics of CRISPR-Cas screens:
(a-b) shows the key steps in performing a CRISPR screen in mammalian cells. Initially the sgRNA library is ordered as a pooled tube of DNA oligonucleotides, typically synthesized commercially via chip based DNA synthesis. The library is then amplified via PCR and cloned into an appropriate lentiviral vector, insuring library coverage is maintained throughout. If the library is obtained in plasmid form (ex. pooled sgRNA libraries available from Addgene), the library simply needs to be transformed into bacteria, expanded, and sequenced to confirm sgRNA representation. Once the library is in a suitable lentiviral vector, the next step is packaging the DNA into lentivirus. Standard lentiviral packaging protocols will suffice, so long as coverage is maintained throughout the packaging. After packaging the lentivirus, a test transduction should be performed to quantify the functional titer (i.e. the actual number of cells transduced per lentiviral particle delivered). This can then be used to determine the amount of lentivirus needed to achieve an MOI of 20-60%. The transduced cells are then passaged with at least 500-1000 fold coverage of the library at each step to ensure accurate sgRNA quantitation. As the cells are passaged, it also is beneficial to store freeze and store aliquots of the library for subsequent massively parallel sequencing. At the end of the functional assay, the library is sequenced a final time to determine the relative enrichment and depletion of specific sgRNA, corresponding to target gene fitness. (c-d) Maintaining library coverage throughout the protocol is essential for insuring statistical confidence and preventing arbitrary library skewing. However, maintaining high coverage of the library imposes significant practical challenges for researchers attempting to implement a CRISPR-Cas screen. The figures above highlight the technical challenges of large library screening, and can serve as a reference for future screen design (bar plots calculated assuming 500 fold coverage of the library). As the number of sgRNA in the library increases, the scale of the experiment may outpace available resources and become untenable. Correspondingly, when planning a CRISPR-Cas genetic screen it is important to determine if the screen is executable in terms of lab equipment, reagents, and manpower. Once the screen has been started, the same mindfulness needs to be directed at insuring there are no library bottlenecking points which could artificially influence the results of the assay.