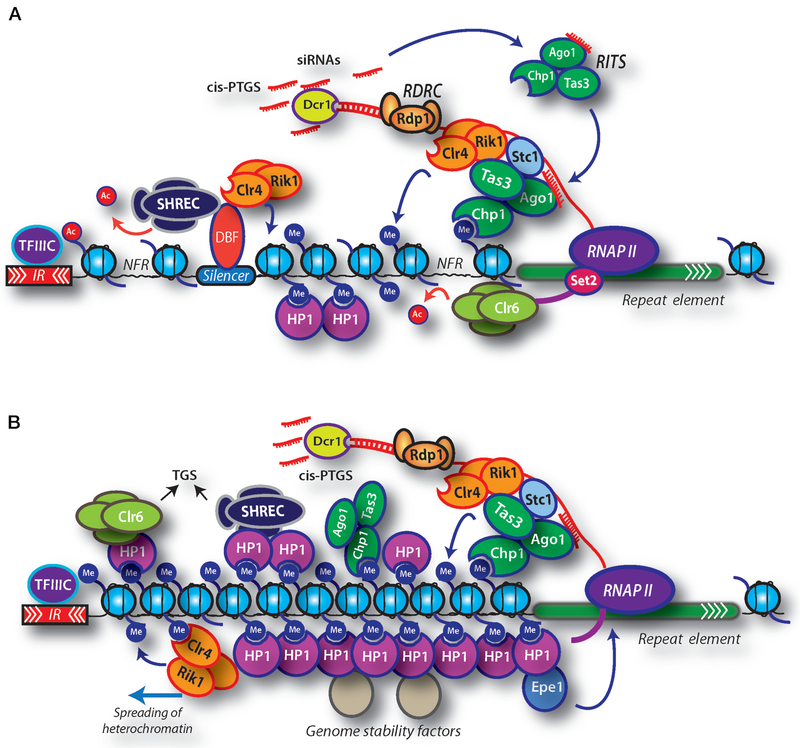
Figure 1.
Heterochromatin assembly in S. pombe. (A) Heterochromatin assembly in S. pombe involves RNAi factors, heterochromatin HP1 proteins (Chp2 and Swi6), and histone-modifying factors including methyltransferase Clr4 and HDAC complexes (SHREC and Clr6). Dicer (Dcr1), RNA-induced transcriptional silencing (RITS) complex, and the RNA-dependent RNA polymerase complex (RDRC) process repeat transcripts into siRNAs that are believed to serve as specificity determinants for the loading of heterochromatin. RNAPII transcription of repeats occurs preferentially during S phase of the cell cycle and is functionally coupled to the loading of Clr4 complex subunit Rik1 and the RITS subunit Ago1 as well as methylation of H3K36 by Set2, which is believed to be important for the loading of Clr6 HDAC. The LIM-domain protein Stc1 facilitates the interaction between Clr4 complex and RITS, which stabilizes their binding to chromatin and promotes the processing of repeat RNAs. Recruitment of Clr4 and HDACs likely involves additional transcription-coupled and noncoding-RNA-coupled mechanisms that act independently of RNAi. In addition, DNA-binding factors (DBF) that recognize specific silencer elements within heterochromatin domains target factors required for heterochromatin assembly. Methylation of histone H3 lysine 9 by Clr4 creates binding sites not only for HP1 proteins but also for the chromodomain of Chp1, a subunit of RITS. The binding of Chp1 to methylated H3K9 (H3K9me) generates a self-reinforcing loop that enables RITS and associated factors to act in cis to process repeat transcripts into siRNAs (cis-PTGS). (B) HP1 proteins bound to H3K9me facilitate the loading of HDACs including SHREC and Clr6. Clr4 is also able to recognize H3K9me via its chromodomain, which is critical for the spreading of heterochromatin. Spreading also requires HP1 proteins and SHREC and involves propagation of heterochromatin-associated modification patterns beyond the initial nucleation sites. The inverted repeat (IR) elements or tRNAs, which are bound by transcription factor TFIIIC (Noma et al. 2006), mark the boundaries of heterochromatin domains and prevent inappropriate spreading of heterochromatin outside of the naturally silenced domains. Chp2- and Swi6-mediated recruitment of SHREC and Clr6 activities is also critical for transcriptional gene silencing (TGS). Clr3 HDAC and Mit1 Snf2 ATPase activities of SHREC facilitate proper positioning of nucleosomes to eliminate nucleosome-free regions (NFRs) within heterochromatin and promote TGS by inhibiting RNAPII occupancy. Swi6 can also recruit an antisilencing factor, Epe1, that stimulates transcription of centromeric repeats in the context of heterochromatin. The assembled heterochromatin structure is stably inherited as an epigenetic state and provides a versatile recruitment platform for various factors that have critical roles in genome integrity and organization.