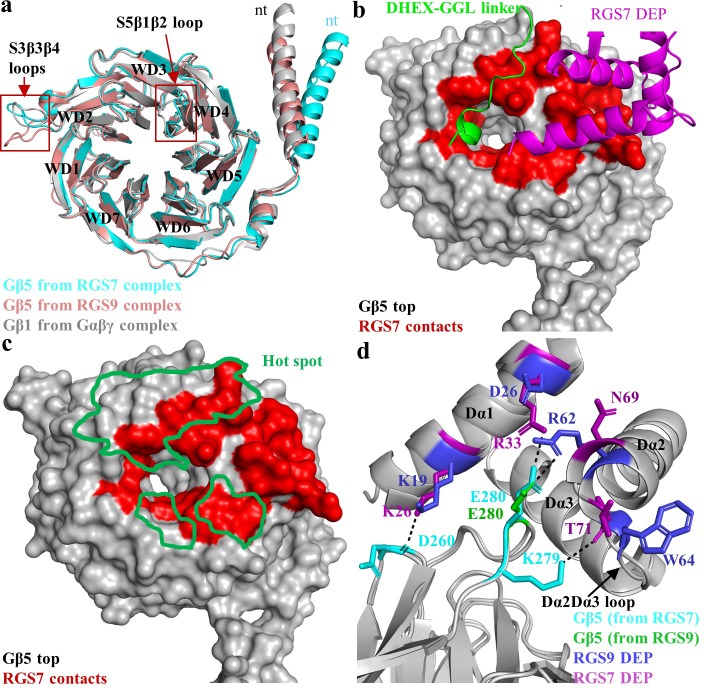
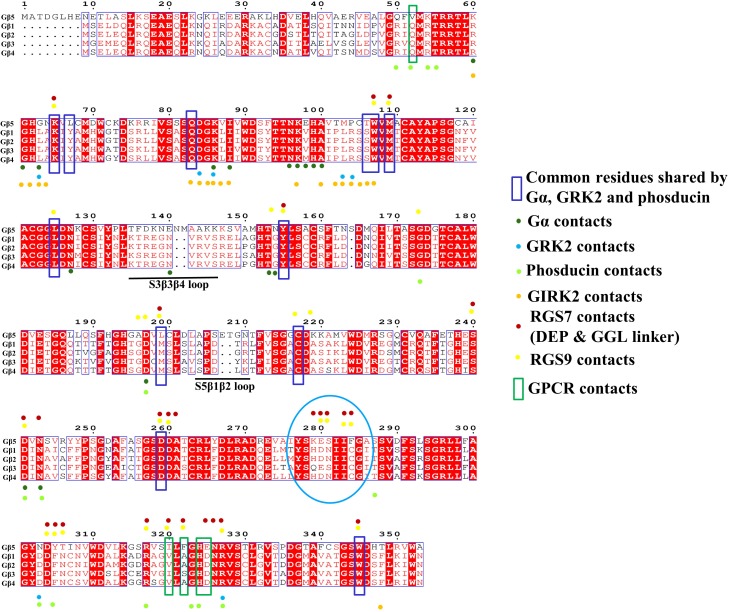
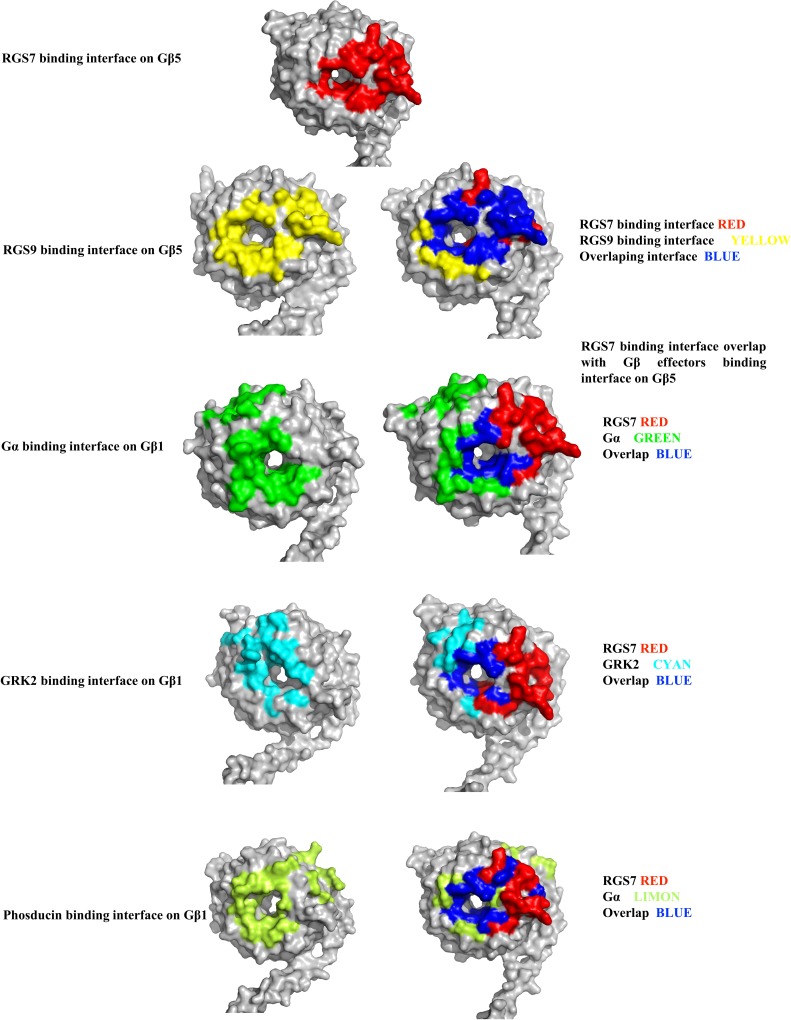
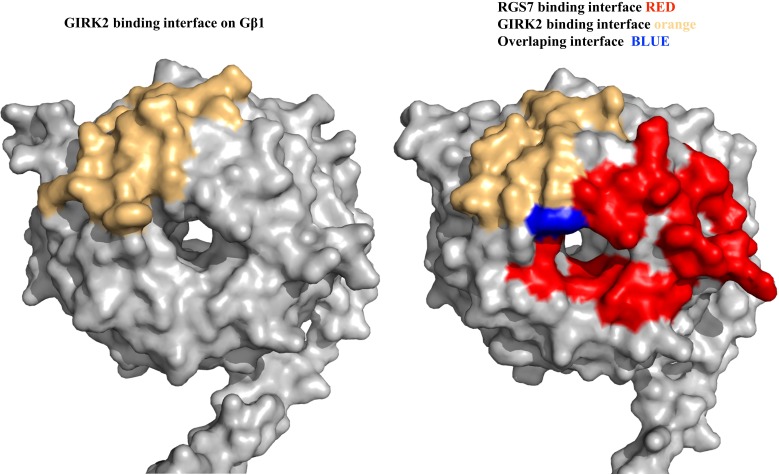
Figure 3. Interaction of RGS7 with Gβ5.
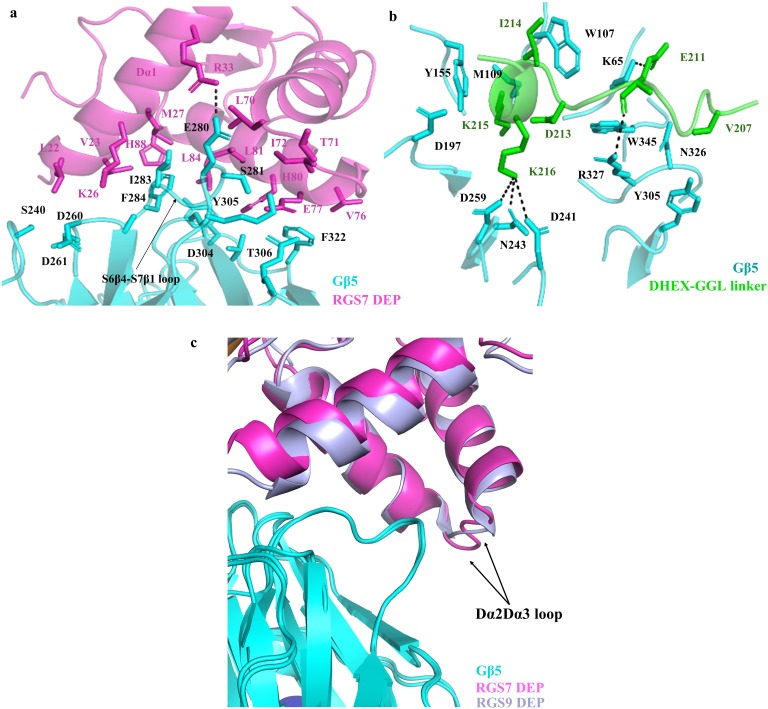
(a) Cartoon representation of the superposed Gβ5 structures from RGS7 with RGS9 and Gβ1. The differences between the loop regions are highlighted in red boxes. The N-terminal α-helix of Gβ5 from RGS7 also shows variation compared to Gβ5 from the RGS9 complex and Gβ1 from the Gαβγ complex. (b) Surface representation of Gβ5 top face (grey) highlighting the RGS7 contact residues (red). Two contact interfaces that are formed by the DHEX-GGL linker and the DEP domain are shown as a carton in green and pink, respectively. (c) The hotspot region (green) mapped onto the Gβ5 surface based on contacts formed by known Gβ1 binding partners. RGS7 (red) overlaps with the hotspot footprint. The DEP domain forms a unique contact surface on the top face of Gβ5. (d) Distinct interacting residues of the DEP domain of RGS7 (pink) that forms direct contacts with Gβ5 (cyan) are shown along with equivalent residues of RGS9. A unique electrostatic interaction is formed by Dα1 residue R33 of the RGS7 DEP domain with E280. Its organizational equivalent in RGS9 is formed by interactions with R62 from α-helix Dα2. The Dα2-Dα3 loop of the DEP domain that is involved in interactions with Gβ5 has a different organization in RGS7 and RGS9 and is indicated by an arrow.