Figure 1.
Cryo-EM Image Reconstruction of MDA5-dsRNA Filaments with Helical Symmetry Averaging
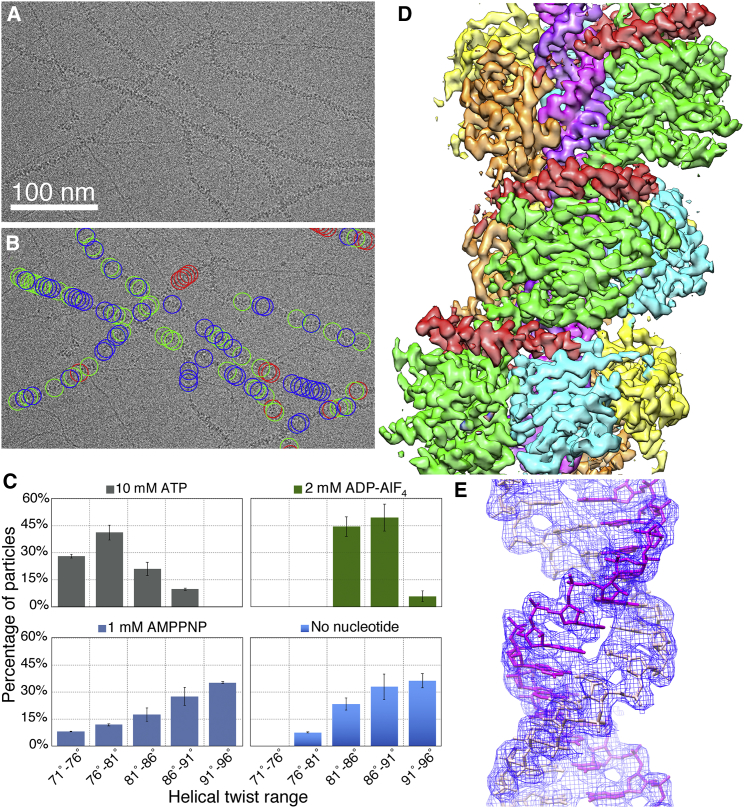
(A) Representative cryo-EM micrograph of MDA5-dsRNA filaments.
(B) Cryo-EM micrograph shown in (A) with circles drawn around the boxed filament segments that were used in the helical reconstructions. The circles are colored according to the 3D class that they contributed to. Segments that contributed to the Twist74, Twist87, and Twist91 structures are in red, green, and blue, respectively.
(C) Histogram showing the distributions of filament segments as a function of helical twist for the ATP, ADP-AlF4, 1-mM AMPPNP, and nucleotide-free datasets. The distributions shown are from 3D classification performed with ten classes per dataset. Error bars represent SEM between replicate 3D classification calculations; n = 3.
(D) 3D density map of the Twist74 MDA5-dsRNA filament at 3.68 Å overall resolution. The components are colored as follows: Hel1, green; Hel2, cyan; Hel2i, yellow; pincer domain, red; CTD, orange; and RNA, magenta.
(E) The dsRNA density in the Twist74 filament (blue mesh) is shown with the fitted atomic model (magenta and pink).
See also Figures S1 and S2.