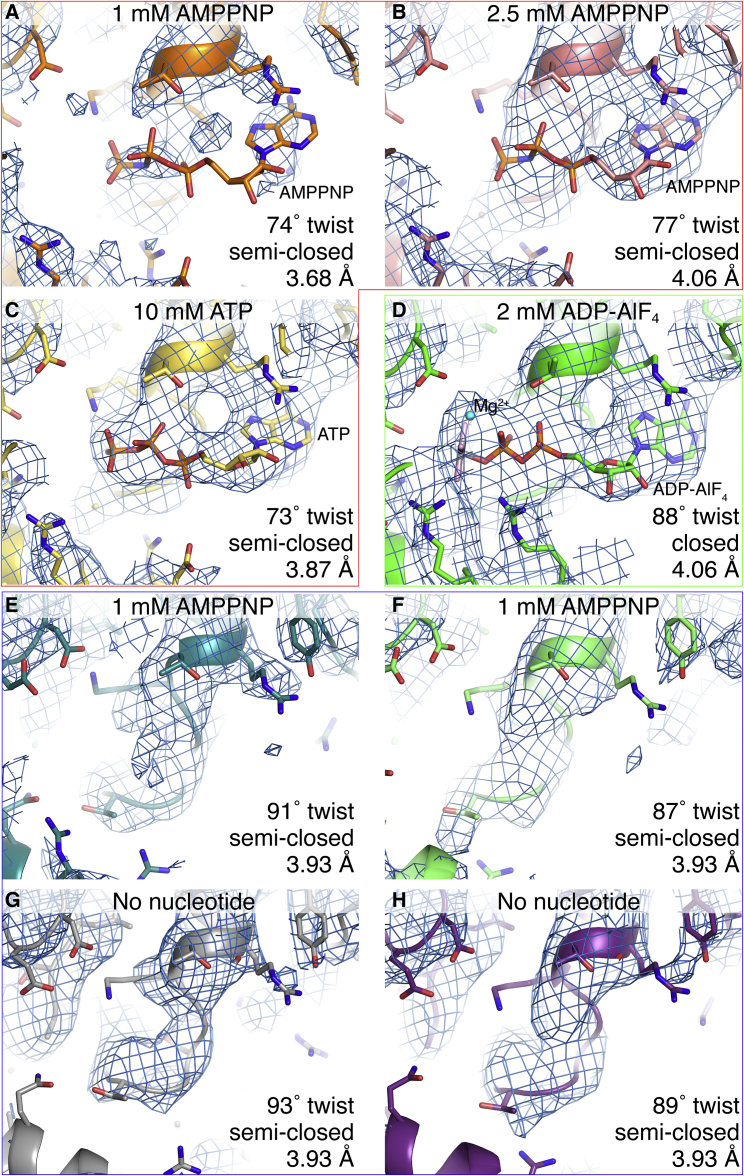
Figure 3.
Close-up Views of the Cryo-EM Densities and Atomic Models around the ATP-Binding Site for Reconstructions with Different Helical Twists and Bound Nucleotides
(A–C) Density consistent with a nucleotide triphosphate molecule is visible in the low-twist structures with 2.5 mM AMPPNP (B) and 10 mM ATP (C), but only weak density is visible in the low-twist (74°) 1 mM AMPPNP structure (A) (red outline).
(D) With 2 mM ADP-AlF4, strong density is visible for the ADP and AlF4 moieties (green box). The AlF4 moiety shown in pink and gray and a coordinated Mg2+ ion in cyan.
(E–H) With 1–2.5 mM AMPPNP (E and F) or no nucleotide (G and H), there is no nucleotide density in the catalytic site of the structures with mid- to high helical twist (81°–96°, blue box).
A contour level of 4.5 σ in PyMol was used for all panels. The AMPPNP, ATP, and ADP-AlF4 molecules and selected protein side chains are shown in stick representation. See also Figure S3.