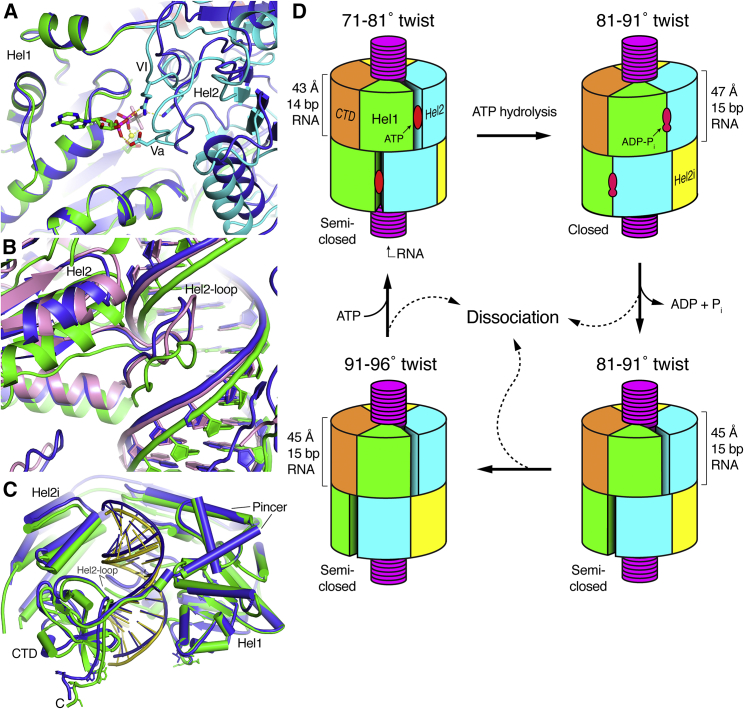
Figure 7.
Comparison of the Closed ADP-AlF4-Bound Structure with the Semi-open Structures and Schematic Model of the ATPase Cycle and Proofreading Mechanism of MDA5
For a Figure360 author presentation of Figure 7, see https://doi.org/10.1016/j.molcel.2018.10.012.
(A) Close-up view of the nucleotide-binding site and Hel1-Hel2 domain interface. The Twist74 AMPPNP-bound structure (blue) was superimposed on the ADP-AlF4-bound structure (colored by domain as in Figure 2) using the Hel1 domain as the reference. Nucleotide-binding motifs Va and VI are labeled. Only the ADP-AlF4 nucleotide is shown for clarity.
(B) Close-up view of the Hel2-loop and its interactions with the dsRNA. The Twist74 (blue) and Twist87 (pink) AMPPNP-bound structures are superimposed onto the ADP-AlF4-bound structure (green) using Hel1 as the reference.
(C) Overview of Twist74 (blue) superimposed on the ADP-AlF4-bound structure (green) using Hel1 as the reference.
(D) Model of the ATPase cycle and proofreading mechanism. Only two filament protomers are shown for clarity. The low-twist (71°–81°) structures correspond to the ATP-bound catalytic ground state, the intermediate-twist (81°–91°) ADP-AlF4-bound structure is the transition state, and the intermediate- and high-twist (91°–96°) states represent nucleotide-free states. The four panels relate to the panels in Figures 3C–3F.
See also Videos S1, S2, S3, S4, and S5.