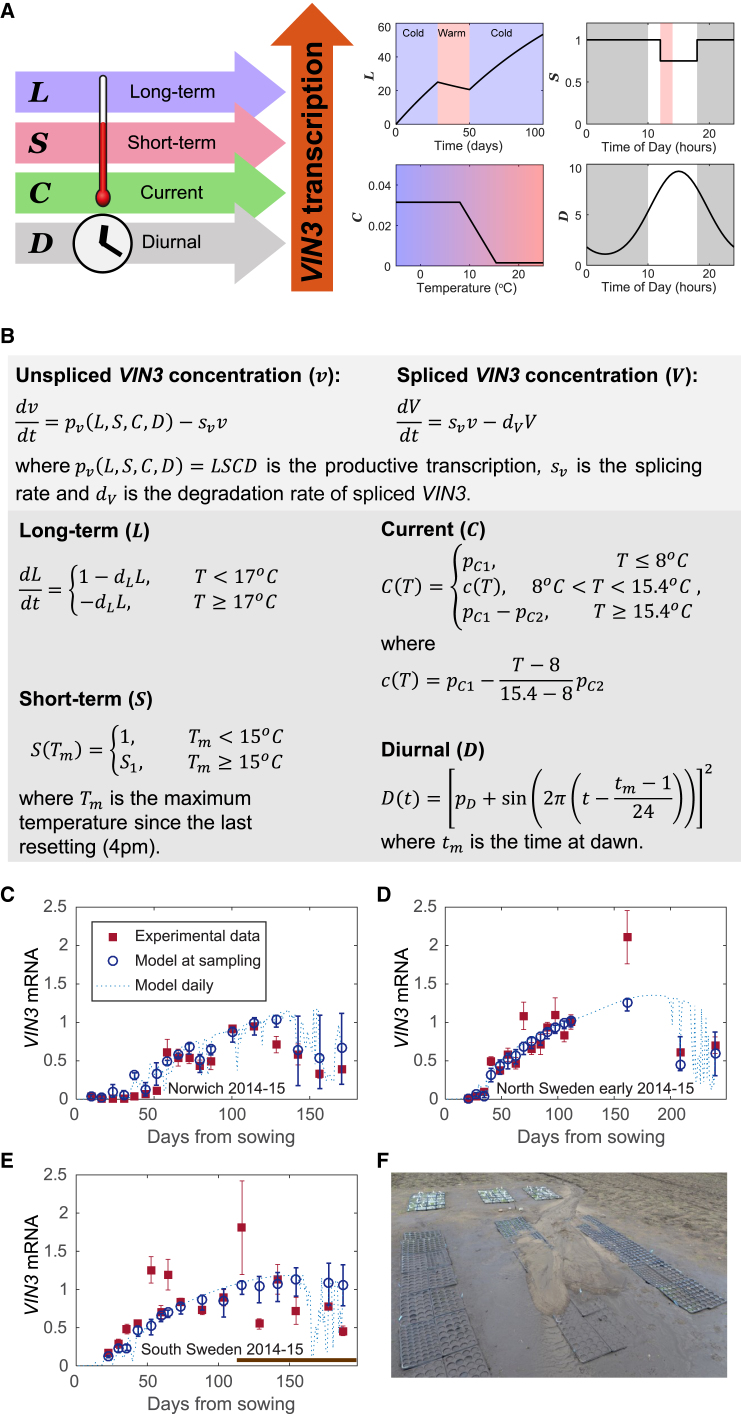
Figure 3.
Description and Fitting of LSCD Model for VIN3 Dynamics
(A) Diagram of the LSCD model showing the primary signals registered by each component, their temperature dependence, and how they affect VIN3 transcription. Element L increases slowly in the cold (<17°C) and decreases slowly in the warm. Element S remembers the presence of a high temperature spike until the evening and, during that time, remains decreased. Element C is high at low temperatures and low at high temperatures, changing linearly with temperature between 8°C and 15.4°C. Element D cycles each day, peaking in the afternoon.
(B) Mathematical description of LSCD model showing the temperature and time dependency of each component.
(C) Comparison of LSCD model and fitted experimental VIN3 mRNA data for Norwich in 2014–2015. Data from Hepworth et al. (2018), bars show mean and standard error, respectively. Model at sampling shows the mean of the predicted values of VIN3 mRNA in the sampling time window, which is defined as the period from 2 hr before the recorded sampling time to 2 hr after due to the long duration of sampling. The error bars show the maximum and minimum values of VIN3 mRNA during that time window. Model daily shows the predicted value for VIN3 mRNA at the same time every day (chosen as the time of the final sampling) to demonstrate the changes in amplitude of the VIN3 daily peak.
(D) Comparison of model and experimental data from North Sweden (early planting) in 2014–2015, as described for Norwich in (C).
(E) Comparison of model and experimental data from South Sweden in 2014–2015, as described for Norwich in (C). The late time points of the South Swedish data (brown bar) could not be fitted by our model, likely due to a mudslide (time given by start of brown bar) that damaged the plants and affected their VIN3 expression.
(F) Mudslide at the South Swedish site covered the plants and caused sample losses.
See also Figures S1–S3 and S5–S7.