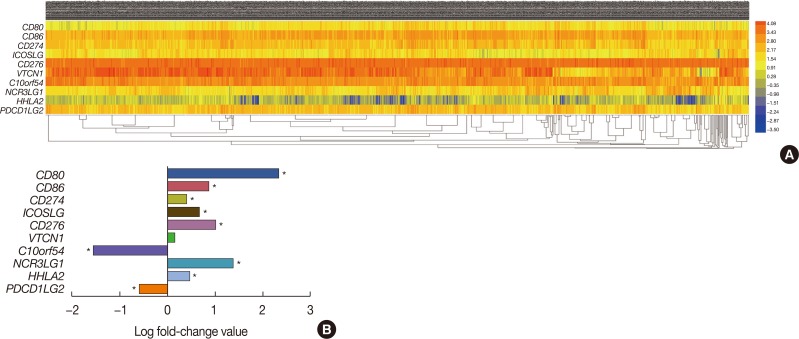
Figure 1. Identification of differentially expressed messenger RNAs (mRNAs) in the The Cancer Genome Atlas (TCGA) BRCA database. (A) Heat map of the log2-fold expression changes in 10 B7 family members mRNAs in the TCGA BRCA database (n=1,092). The horizontal row represents different mRNAs, and the vertical column represents different patients. Green squares indicate increases, and red squares indicate decreases. (B) Statistical analysis of altered expression of B7 family members in the TCGA BRCA data. The false discovery rate for all data is less than 0.01.
CD80=cluster of differentiation 80; CD86=cluster of differentiation 86; CD274=cluster of differentiation 274; ICOSLG=inducible T-cell costimulator ligand; CD276=cluster of differentiation 276; VTCN1=V-set domain containing T cell activation inhibitor 1; C10orf54=chromosome 10 open reading frame 54; NCR3LG1=natural killer cell cytotoxicity receptor 3 ligand 1; HHLA2=HERV-H LTR-associating 2; PDCD1LG2=programmed cell death 1 ligand 2. *p<0.05.