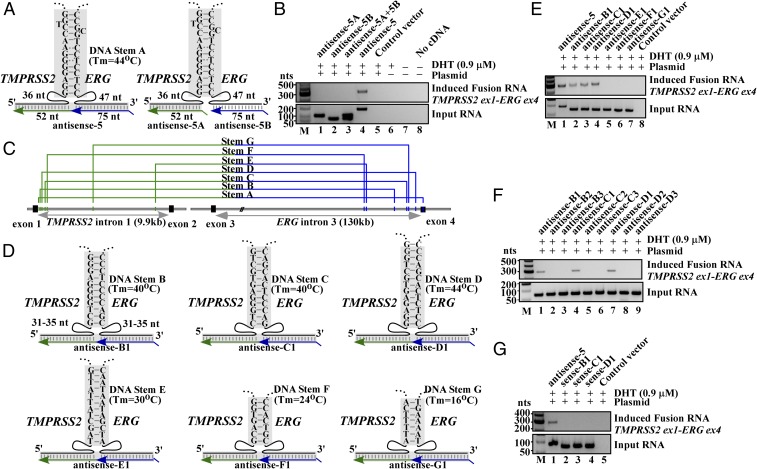
Fig. 2.
Formation of a three-way junction may facilitate fusion induction. (A, Left) Schematics of three-way junction that could be formed between genomic DNA (black) and antisense-5 input RNA (green/blue). The sense genomic strands of both TMPRSS2 and ERG genes are on the minus strand of chromosome 21, separated by 3 Mb. Short lines in shaded regions represent base pairings. Imperfect DNA stem A includes a high-energy G-T and A-C wobble pair known to have Watson–Crick-like geometry in a DNA double helix (30, 31). A spacer region of 36 nt and 47 nt separate stem A from the regions targeted by antisense-5 input RNA. (Right) Expressing antisense-5 as two separate halves (antisense-5A and -5B) that severed the link between TMPRSS2 (52 nt) and ERG (75 nt) RNA sequence. (B) RT-PCR assays of fusion transcripts showed that the severed input RNAs resulted in the loss of fusion transcript induction (lanes 1, 2, 3, vs. lane 4). (C) Locations of putative stems A to G identified by BLAST analyses. The DNA stem B to G are located in the introns. Genomic coordinates are listed in SI Appendix, Fig. S9. (D) The putative three-way junction formed between the indicated genomic DNA stem B to G (black) and designed antisense input RNA (green/blue). These antisense RNAs target introns and contain no exon sequence. (E) Targeting genomic DNA stem B, C, and D that exhibit higher DNA stem stability (Tm = 40 °C, 40 °C, and 44 °C, respectively) by antisense RNAs induced fusion transcripts (lanes 2–4). In contrast, targeting less stable stem E, F, and G (Tm = 30 °C, 24 °C, and 16 °C, respectively) failed to induce fusion transcripts (lanes 5–7). (F) Antisense input RNAs designed to invade each of the respective genomic DNA stem B, C, and D (antisense-B2, B3, C2, C3, D2, and D3) resulted in the loss of induction. (G) Corresponding sense input RNAs targeting stem B, C, and D failed to induce fusion transcripts (lanes 2–4).