Fig. 3.
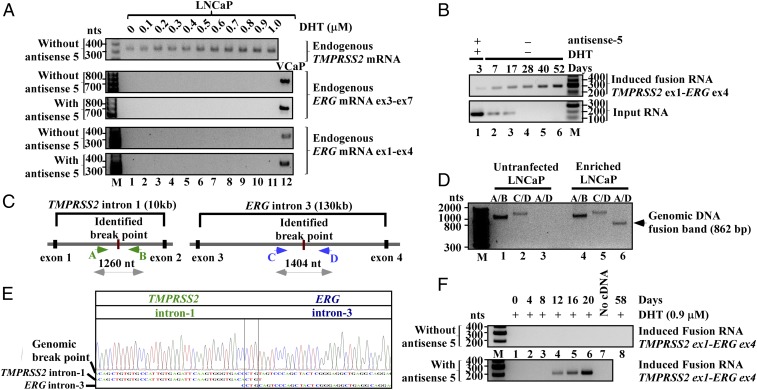
Induced TMPRSS2–ERG fusion is the result of genomic arrangements. (A, Top) RT-PCR shows that LNCaP cells express TMPRSS2 mRNA, which is up-regulated by DHT. Primers used are specific to TMPRSS2 exon-2 and exon-4. (Middle and Bottom) ERG mRNA, however, was not detected in LNCaP cells under a wide range of DHT in the presence or absence of antisense-5 (lanes 1–11). RT-PCR assays were performed using two independent primer pairs that selectively amplify exon-3 to -7 (Middle), or exon-1 to -4 (Bottom) of ERG mRNA. Both primer pairs amplified ERG mRNA in VCaP cells (lane 12), but will not amplify the induced TMPRSS2–ERG fusion transcript which has ERG exon-4 to -12. (B) RT-PCR shows the transient nature of input RNA that was degraded by day 17 (Lower), and the persistent nature of the induced fusion transcript (Upper) up to 52 d postinitial treatment in the enriched LNCaP population (see SI Appendix, Fig. S14 for enrichment procedure). (C) Schematics of identified genomic breakpoints and the primer A, B, C, and D used to amplify the breakpoints. (D) The unrearranged wild-type TMPRSS2 and ERG alleles were revealed by primer pair A/B (∼1,404 bp) and C/D (∼1,260 bp), respectively (lanes 1, 2, 4, and 5). The genomic fusion band of 862-bp amplified by fusion-specific primer pair A/D was present only in the enriched LNCaP population (lane 6) and absent in untransfected LNCaP cells (lane 3). (E) Sanger sequencing of the fusion band showed a 500-bp segment of TMPRSS2 intron-1 fused to 362 bp of ERG intron-3 defined by primer A/D. The genomic breakpoint contains a “CTG” microhomology (boxed). The full-length Sanger sequence is shown in SI Appendix, Fig. S16. (F) Prolonged expression of antisense-5 for 12 d induced the TMPRSS2–ERG fusion transcript in PNT1A cells as detected by three-round nested PCR.