Significance
In the proteasome, an essential protease of eukaryotes, the hexameric ATPases (Rpt1-Rpt6) inject ubiquitinated proteins into the proteolytic core particle. Individual Rpt proteins assemble into the hexameric Rpt ring through binding to specific chaperones: Nas2, Hsm3, Rpn14, and Nas6. Here, we show that Rpt ring assembly uses a ubiquitination-mediated control. Not4 ubiquitinates Rpt5 by competing against Nas2 first and then Hsm3 throughout Rpt ring assembly. Thus, Not4 selectively recognizes a Rpt ring that matures without these chaperones. Rpt5 ubiquitination blocks recruitment of Rpn1 ubiquitin receptor and Ubp6 deubiquitinase, halting proteasome assembly and its recognition of ubiquitinated substrates. Our findings reveal an assembly checkpoint where Not4 monitors chaperone actions during hexameric ATPase ring assembly, ensuring the accuracy of proteasome holoenzyme maturation.
Keywords: proteasome, assembly chaperone, AAA+ ATPase, checkpoint, Not4
Abstract
In the proteasome holoenzyme, the hexameric ATPases (Rpt1-Rpt6) enable degradation of ubiquitinated proteins by unfolding and translocating them into the proteolytic core particle. During early-stage proteasome assembly, individual Rpt proteins assemble into the hexameric “Rpt ring” through binding to their cognate chaperones: Nas2, Hsm3, Nas6, and Rpn14. Here, we show that Rpt ring assembly employs a specific ubiquitination-mediated control. An E3 ligase, Not4, selectively ubiquitinates Rpt5 during Rpt ring assembly. To access Rpt5, Not4 competes with Nas2 until the penultimate step and then with Hsm3 at the final step of Rpt ring completion. Using the known Rpt–chaperone cocrystal structures, we show that Not4-mediated ubiquitination sites in Rpt5 are obstructed by Nas2 and Hsm3. Thus, Not4 can distinguish a Rpt ring that matures without these chaperones, based on its accessibility to Rpt5. Rpt5 ubiquitination does not destabilize the ring but hinders incorporation of incoming subunits—Rpn1 ubiquitin receptor and Ubp6 deubiquitinase—thereby blocking progression of proteasome assembly and ubiquitin regeneration from proteasome substrates. Our findings reveal an assembly checkpoint where Not4 monitors chaperone actions during hexameric ATPase ring assembly, thereby ensuring the accuracy of proteasome holoenzyme maturation.
The proteasome holoenzyme is a complex molecular machine responsible for ubiquitin-mediated protein degradation (1). The proteasome holoenzyme forms via the association between the 28-subunit core particle (CP) and 19-subunit regulatory particle (RP) (2–5). The RP recognizes polyubiquitinated proteins and translocates them in an ATP-dependent manner into the CP, where protein degradation occurs (1). The CP is a stack of four heteroheptameric rings (α1–7β1–7β1–7α1–7) (6). The inner β rings contain proteolytic sites, and the outer α rings provide the binding sites for the RP. The RP forms via the joining of two subassemblies: a nine-subunit base and a nine-subunit lid (7). In the RP, a major structural and functional platform exists in the base, in which six ATPase subunits (Rpt1 to Rpt6) are specifically arranged into the heterohexameric Rpt ring. The Rpt ring provides specific binding sites for the lid to form the RP, and the ring also directly associates with the CP for both assembly and function of the proteasome holoenzyme (2, 4, 5, 8, 9).
Four evolutionarily conserved chaperones (Hsm3, Nas2, Rpn14, and Nas6) govern heterohexameric Rpt ring assembly of the base (10–14). These chaperones bind to specific Rpt proteins in a pairwise manner: Rpn14-Rpt6, Nas6-Rpt3, Hsm3-Rpt1, and Nas2-Rpt5 (11, 12, 14). Each chaperone is found in distinct “Rpt-Rpt modules”: Rpn14-Rpt6-Rpt3-Nas6, Rpt4-Rpt5-Nas2, and Hsm3-Rpt1-Rpt2. These Rpt-Rpt modules join to complete the heterohexameric Rpt ring of the base. Although these chaperones are phylogenetically and structurally unrelated, they exhibit a common feature that they sterically block the base’s interactions with the CP (11, 15, 16). Nas6 can also block base–lid interactions (17). These data have established a current model, in which the chaperones cooperatively obstruct premature base–lid and base–CP associations, until the heterohexameric Rpt ring of the base is complete (11, 15–17). The chaperones remain on the base and RP until these complexes properly associate with the CP. Completion of the proteasome holoenzyme (lid–base–CP) results in an eviction of the chaperones (10, 11, 16–18).
The current model of proteasome assembly illustrates that the base assembles first, before its progression to higher-order complexes. However, there is little mechanistic understanding as to how completion of base assembly might be ensured. Based on structural and biochemical data, the fully formed base is in complex with the chaperones at specific positions (4, 19, 20). Rpn14 and Nas6 are on one side of the hexameric Rpt ring, whereas Nas2 and Hsm3 are on the opposite side, reflecting a specific arrangement of their cognate Rpt proteins. In particular, Nas2 and Hsm3 initially exist in different modules and become direct neighbors only upon completion of the heterohexameric Rpt ring (20, 21). In the ordered assembly of the base, the Nas2 module incorporates at the penultimate step, and the Hsm3 module incorporates at the last step. Although the specific sequence of assembly is well established, it remains unknown whether and how the stepwise progression of base assembly might be monitored to ensure proper proteasome holoenzyme formation.
In the present study, we demonstrate that chaperone-mediated base assembly employs a ubiquitination-dependent control through an E3 ligase, Not4. Not4 ubiquitinates Rpt5 in the nascent base by competing against Nas2 and Hsm3, which sequentially obstruct the ubiquitination sites on Rpt5. Ubiquitin on Rpt5 hinders docking of Ubp6 deubiquitinase and its receptor Rpn1 to the base, thereby stalling base assembly. On the other hand, the fully formed base can readily recruit Rpn1 and Ubp6 and proceed to proteasome holoenzyme formation. Not4-mediated control provides a mechanism to track the last steps of base assembly to ensure completion of the base, before its progression to the fully formed proteasome holoenzyme.
Results
Not4 Ubiquitin Ligase Regulates the Progression of Base Assembly.
Multiple assembly chaperones cooperatively promote heterohexameric ATPase ring assembly of the base for proteasome holoenzyme formation (10–14, 16, 22, 23). Although interactions between the chaperones and their cognate ATPases provide a major mode of regulation during base assembly, genetic and biochemical data suggest an additional mode of regulation involving ubiquitination (24, 25). In particular, deletion of NOT4, which encodes an E3 ubiquitin ligase, has been suggested to influence structural features of the base and its association in the proteasome holoenzymes (25). Because Not4 is neither a component nor a regulator of the proteasome holoenzyme, we examined whether Not4 affects chaperone-mediated assembly of the base through its E3 activity. Because the not4Δ mutant exhibits pleiotropic effects (SI Appendix, Fig. S1), we specifically disabled the E3 ligase activity of Not4 by generating a L35A substitution; this mutation is known to abrogate Not4’s interaction with its cognate E2 enzyme (26). We then combined the not4-L35A allele with a panel of yeast strains lacking the chaperones, Rpn14, Nas6, Hsm3, and Nas2, in combinations (Fig. 1A).
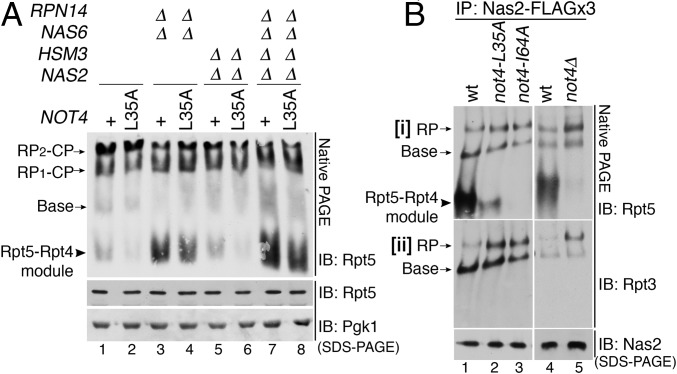
Fig. 1.
Not4 ubiquitin ligase regulates the progression of base assembly. (A) The not4-L35A catalytic mutants affect base assembly, as reflected in the Rpt5-Rpt4 module. Whole-cell extracts (80 μg) were analyzed by 3.5% native PAGE and immunoblotting for Rpt5, a subunit of the base. The not4-L35A allele is integrated into the chromosomal locus of NOT4. The plus (+) indicates the wild-type NOT4. Pgk1, loading control. (B) The not4 catalytic mutants exhibit enhanced progression of base assembly, as indicated by increased RP (base–lid) relative to the Rpt5-Rpt4 module. Assembly intermediates were affinity-purified via 3× FLAG-tagged Nas2 and were analyzed by 3.5% native PAGE and immunoblotting for the indicated Rpt proteins (i and ii). Nas2 serves as a loading control.
We first examined relative levels of assembly intermediates and the proteasome holoenzyme by analyzing whole-cell extracts through native PAGE and immunoblotting with Rpt5, a representative ATPase subunit of the base. In wild-type cells, the Rpt5-Rpt4 module, an early assembly intermediate, is scarce relative to the proteasome holoenzyme (RP2-CP, RP1-CP), indicating its robust assembly into the proteasome holoenzyme (Fig. 1A, lane 1). In contrast, in the chaperone-deletion mutants, the Rpt5-Rpt4 module accumulates with an accompanying decrease in proteasome holoenzyme levels, indicating inefficient assembly into the proteasome holoenzyme (Fig. 1A, lanes 3 and 7) (10, 12, 13, 17). Unexpectedly, the Rpt5-Rpt4 module is decreased in the not4-L35A mutants relative to wild type (Fig. 1A, compare lanes 2 and 1). This trend is also observed when the not4-L35A was combined with the chaperone-deletion mutants (Fig. 1A, compare lanes 4 and 3, 6 and 5, and 8 and 7). The total cellular level of Rpt5 is comparable in all samples (Fig. 1A, Middle), indicating that Not4 does not affect stability of Rpt5.
Two alternative models could explain the reduction of the Rpt5-Rpt4 module in the not4-L35A mutants in Fig. 1A. First, the Rpt5-Rpt4 module may not form efficiently. Second, this module could form and readily proceed to higher-order complexes, thereby resulting in its depletion. The first model predicts a decrease in late intermediates, base and RP, whereas the second model predicts an increase in these complexes. To distinguish between these two models, we tracked Rpt5 from early- to late-stage RP assembly by isolating Rpt5-containing complexes via its cognate Nas2 chaperone, harboring a 3× FLAG affinity tag in its chromosomal locus. In wild-type cells, Nas2-bound complexes yielded mainly the Rpt5-Rpt4 module and only some base and RP (Fig. 1 B, i, lanes 1 and 4). In contrast, in not4-L35A, not4-I64A, and not4Δ, Nas2-bound complexes mainly yielded base and RP and almost no or little detectable Rpt5-Rpt4 module (Fig. 1 B, i, lanes 2, 3, and 5); the I64A substitution is equivalent to the L35A substitution (26). Using mass spectrometry, we confirmed that all bona fide subunits exist in the base and RP (SI Appendix, Tables S1 and S2). Overall, RP is increased in the not4 mutants relative to wild type (Fig. 1 B, ii, lanes 2, 3, and 5). Our data support the second model, in which the Rpt5-Rpt4 module readily progresses into the base and RP in the not4 mutants, suggesting that Not4 E3 ligase provides a negative regulatory role during base assembly.
Not4 E3 Ligase Plays an Antagonistic Role During Chaperone-Mediated Proteasome Assembly.
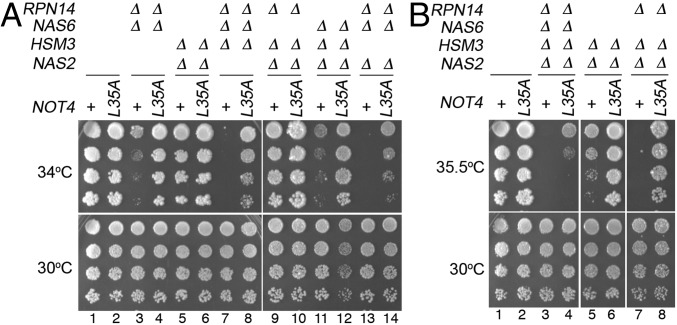
To understand the in vivo function of Not4’s ubiquitinating activity during proteasome assembly, we examined growth phenotypes of the chaperone-deletion mutants harboring the not4-L35A mutant. The not4-L35A mutant alone grows indistinguishably from wild type (Fig. 2A, lanes 1 and 2). As shown previously, cells lacking multiple chaperones exhibit severe growth defects even upon mild heat stress at 34 °C, reflecting deregulated proteolysis due to defective proteasome assembly (Fig. 2A, Top, lanes 3, 7, 11, and 13) (11–13). Remarkably, when the not4-L35A allele is expressed in these strains, it potently restores growth of these chaperone-deletion mutants at 34 °C (Fig. 2A, Top, lanes 4, 8, 12, and 14) and also at 35.5 °C for two mutants, nas2Δhsm3Δ and nas2Δhsm3Δrpn14Δ (Fig. 2B, Top, lanes 6 and 8). Similar to the not4-L35A allele, the not4-I64A allele also restores growth of the chaperone mutants (SI Appendix, Fig. S2). Although the quadruple chaperone knockout cells die at 35.5 °C, they show some survival when combined with either the not4-L35A or not4-I64A allele (Fig. 2B, Top, compare lanes 3 and 4; and SI Appendix, Fig. S2B). Also, these growth phenotypes reflect the specific ubiquitin ligase function of Not4, rather than the general function of the Ccr4-Not complex (SI Appendix, Fig. S3). Together, these data demonstrate that Not4 E3 ligase profoundly influences the entire chaperone-mediated base assembly process.
Fig. 2.
Not4 ubiquitin ligase provides inhibitory control during base assembly, as indicated by phenotypic suppression of the chaperone mutants by the not4 mutants. (A and B) Growth assays showing that the not4-L35A allele restores the growth of the chaperone-deletion mutants upon heat stress (Top). Threefold serial dilutions of indicated yeast cells were spotted onto yeast extract–peptone–dextrose plates and grown at the indicated temperature for 2–4 d.
In summary, our genetic data show that the not4 mutant restores growth of the chaperone-deletion mutants, suggesting that Not4 provides an inhibitory control during chaperone-mediated base assembly, in line with our data in Fig. 1. Because Not4 function is strongly detected upon loss of the chaperones, Not4’s ubiquitination targets are not the chaperones but are likely to be their binding partners, namely, Rpt proteins. This relationship suggests that Not4 may act on Rpt proteins in competition against the chaperones during proteasome assembly.
Not4 Ubiquitinates Rpt5, Acting in Competition Against the Chaperones Nas2 and Hsm3.
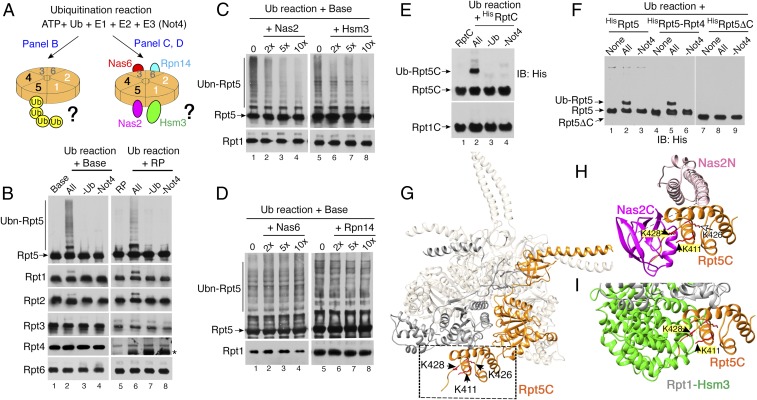
Because our genetic data suggest a model that Not4 may ubiquitinate Rpt proteins upon loss of the chaperones (Fig. 2), we examined whether Not4 directly ubiquitinates any Rpt subunit. We established an in vitro ubiquitination assay using ATP, ubiquitin, and an E1 (Uba1), E2 (Ubc4), and E3 (Not4) (Fig. 3A) (26). We then individually added two well-established assembly intermediates, the base and RP, in which all six Rpt proteins are present. Following the ubiquitination reactions, we examined whether any Rpt subunit exhibits high-molecular-weight conjugates, representing ubiquitinated Rpt species. Out of six Rpt subunits, Rpt5 uniquely exhibited both monoubiquitinated and polyubiquitinated species (Fig. 3B, lanes 2 and 6), which were not detected when either ubiquitin or Not4 was omitted (Fig. 3B, lanes 3, 4, 7, and 8). Other Rpt subunits were not strongly ubiquitinated, although all six Rpt subunits have a similar number of surface lysines as potential ubiquitination sites (Fig. 3B and SI Appendix, Figs. S4 and S5). Together, our data suggest that Rpt5 is a major ubiquitination target of Not4 and that Rpt5 can be ubiquitinated following its assembly into the base and RP complexes.
Fig. 3.
Not4 ubiquitinates Rpt5 by competing with both Nas2 and Hsm3 in the base. (A) An experimental scheme for in vitro ubiquitination reactions. The heterohexameric Rpt ring is shown with specific positioning of individual Rpt subunits (Left) and their cognate chaperones (Right) (4, 20). (B) Not4 ubiquitinates Rpt5 in the base and RP. The base and RP (2 pmol each) were subjected to ubiquitination reactions and then analyzed by 10% Bis-Tris SDS/PAGE and immunoblotting for each Rpt subunit. Asterisk, nonspecific signal (SI Appendix, Fig. S5). (C and D) Nas2 and Hsm3 block Not4-mediated ubiquitination of Rpt5, whereas Nas6 and Rpn14 do not. Each chaperone was added at the indicated molar excess over the base (2 pmol) during ubiquitination reactions and then analyzed as in B. Rpt1, a loading control. (E and F) Not4 ubiquitinates the Rpt5 C-domain (Rpt5C). Ubiquitination reactions were conducted using the recombinant Rpt5C, full-length Rpt5, Rpt5-Rpt4 cocomplex and Rpt5ΔC lacking the C-domain (75 pmol each). In all cases, Rpt5 is His6-tagged. “None” in F indicates substrates only, without ubiquitination reaction. (G) Rpt5 (orange) in the heterohexameric Rpt ring [Protein Data Bank (PDB) ID code 4CR2] (42); Rpt1, gray; the other Rpt proteins, beige. Rpt5C is indicated by the dotted box. Arrowheads indicate Not4-mediated ubiquitination sites: K411, K426, and K428 (SI Appendix, Fig. S7). G–I were generated by University of California, San Francisco Chimera (43). (H and I) Rpt5 ubiquitination sites (yellow highlights) are obstructed by Nas2 C-domain (magenta, PDB ID code 4O06; SI Appendix, Fig. S8) (21, 44) in H and Hsm3 from Rpt1-Hsm3 cocrystal structure (PDB ID code 4JPO) (21), which was superimposed onto the heterohexameric Rpt ring structure (PDB ID code 4CR2) (42) in I. Rpt1 is shown in gray.
During base assembly, Rpt5 binds to its cognate chaperone, Nas2 (Fig. 3A, Right) (12–14). Therefore, we tested whether this Nas2-Rpt5 interaction affects Not4-mediated ubiquitination of Rpt5. We added Nas2 in an increasing molar ratio to Rpt5 in the base and then conducted ubiquitination reactions. When Nas2 was present during the ubiquitination reactions, Rpt5 ubiquitination was noticeably decreased in a dose-dependent manner (Fig. 3C, lanes 2–4). These data suggest that the Nas2-Rpt5 association blocks Rpt5 ubiquitination by Not4.
In the fully formed heterohexameric Rpt ring, Rpt5 is positioned directly adjacent to Rpt1 and its cognate chaperone, Hsm3 (Fig. 3A, Right) (20). The Hsm3-Rpt1-Rpt2 module is the last to be added to complete the heterohexameric Rpt ring, and this event is accompanied by the release of Nas2 (20, 21). Based on the proximity between Hsm3 and Rpt5 in the heterohexameric Rpt ring, we tested whether Hsm3 could also affect Not4-mediated ubiquitination of Rpt5. When Hsm3 was added in an increasing molar ratio to the base during ubiquitination reactions, Not4-mediated ubiquitination of Rpt5 was decreased in a dose-dependent manner (Fig. 3C, lanes 6–8). In contrast, the other chaperones, Nas6 and Rpn14, which bind to the opposite side from Rpt5 in the base (Fig. 3A, Right), did not block ubiquitination of Rpt5 by Not4 (Fig. 3D). Together, these data suggest that Nas2 and Hsm3 can individually block Not4-driven ubiquitination of Rpt5 during heterohexameric Rpt ring assembly.
Nas2 and Hsm3 Obstruct the Same Ubiquitination Sites on Rpt5.
Because Nas2 binds to Rpt5 in the C-terminal domain (C-domain) during Rpt ring assembly (12, 27), we hypothesized that ubiquitination sites may exist in the Rpt5 C-domain, and that Nas2 binding sterically occludes them from Not4. To test this hypothesis, we expressed and purified His6-tagged Rpt5 C-domain from E. coli. Indeed, Not4 ubiquitinated the Rpt5 C-domain but not the Rpt1 C-domain (Fig. 3E, lane 2), supporting our conclusion that Rpt5 is a major ubiquitination target of Not4 (Fig. 3B). Similarly, Not4 ubiquitinated the full-length Rpt5 but not its truncation mutant lacking the C-domain (Fig. 3F, compare lanes 2 and 8). Not4 also ubiquitinated Rpt5 in the Rpt5-Rpt4 cocomplex (Fig. 3F, lane 5, and SI Appendix, Fig. S6), and this ubiquitination was diminished when Nas2 was added (SI Appendix, Fig. S6B). These results demonstrate that the Rpt5 C-domain provides ubiquitination sites for Not4 and that Rpt5 is mainly monoubiquitinated in an early stage of base assembly and is further polyubiquitinated upon its incorporation into the base.
Using a proteomics approach, we determined that Not4 ubiquitinates three lysines of the Rpt5 C-domain: K411, K426, and K428 residues (Fig. 3G and SI Appendix, Fig. S7). When we substituted these three lysines with alanines, the Rpt5 C-domain was no longer ubiquitinated (SI Appendix, Fig. S7C). To examine whether these lysines are sterically occluded by the chaperone binding, we used structural modeling. In the Nas2-Rpt5 structural model, Nas2 obstructs both K411 and K428 in Rpt5 (Fig. 3H and SI Appendix, Fig. S8). This finding is in line with the previous data showing that Nas2 requires the entire Rpt5 C-domain for the Nas2-Rpt5 association (27). When the known Hsm3-Rpt1 cocrystal structure was superimposed onto the heterohexameric Rpt ring structure, Hsm3 also obstructs the two lysines, K411 and K428, of Rpt5, the same as Nas2 (Fig. 3I). The third lysine, K426 in Rpt5 (Fig. 3H), is obstructed by Nas2 in the ATPγS-bound state model of the heterohexameric Rpt ring (SI Appendix, Fig. S8C), suggesting that access to these lysines may also be regulated in a conformation-dependent manner during base assembly (17, 28). Our structural modeling agrees well with our experimental data and demonstrates that Nas2 and Hsm3 obstruct the same ubiquitination sites on Rpt5 from Not4.
Not4 Ubiquitinates Rpt5 at Sequential Stages During Endogenous Base Assembly.
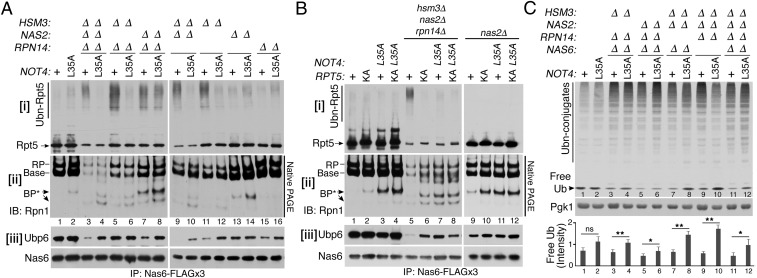
Given that Not4 competes with Nas2 and Hsm3 to access Rpt5 (Fig. 3), we examined whether Not4 ubiquitinates Rpt5 when these chaperones are absent. To track Not4-mediated ubiquitination of Rpt5 in vivo, we isolated assembly intermediates from a panel of chaperone-deletion mutants, using 3× FLAG-tagged Nas6 because it incorporates Rpt modules from the onset of base assembly until its completion (12, 13, 17) and Nas6 does not block Rpt5 ubiquitination (Fig. 3 A and D). In wild-type cells, Rpt5 ubiquitination was not readily detectable in Nas6-bound assembly intermediates (Fig. 4 A, i, lane 1). Strikingly, upon deletion of HSM3 singly or in combination with the other chaperones (Fig. 4 A, i, lanes 3, 5, 9, and 11), high-molecular-weight Rpt5 species were readily detected in the Nas6-purified complexes. These Rpt5 species were undetectable in the corresponding complexes from Not4-L35A–expressing cells (Fig. 4 A, i, lanes 4, 6, 10, and 12), indicating that Not4’s ubiquitinating activity is responsible for the formation of high-molecular-weight Rpt5 species. We confirmed that the high-molecular-weight Rpt5 species indeed represented ubiquitinated Rpt5 by treating them with a deubiquitinase with broad specificity, Usp2, which abolished these species (SI Appendix, Fig. S9) (29). These results demonstrate that Not4 ubiquitinates Rpt5 in the endogenous base species that assemble without Hsm3, supporting the mode of Not4 action as predicted from our in vitro ubiquitination data and structural modeling (Fig. 3).
Fig. 4.
Not4-mediated ubiquitination of Rpt5 blocks recruitment of Ubp6 and Rpn1 during endogenous base assembly. (A and B) Not4-mediated ubiquitination of Rpt5 blocks incorporation of Rpn1 and Ubp6 into the base. Assembly intermediates were isolated using 3× FLAG-tagged Nas6 and were analyzed by 4–12% Bis-Tris SDS/PAGE and immunoblotting for indicated proteins in i and iii (See SI Appendix, Supplementary Materials and Methods for details). Rpn1-containing species were detected by 3.5% native PAGE and immunoblotting (ii). Nas6, a loading control (iii). (C) Not4-mediated control of proteasome assembly regulates the free ubiquitin pool. Whole-cell lysates (20 μg) were subjected to 10% Bis-Tris SDS/PAGE and immunoblotted for ubiquitin (SI Appendix, Supplementary Materials and Methods). Pgk1, loading control. Relative signal intensities of free ubiquitin bands were quantified (n = 4; mean ± SEM; ns, not significant; *P < 0.05; **P < 0.01).
In the nas2Δ single mutants, Rpt5 ubiquitination was not readily detectable (Fig. 4 A, i, lane 13), indicating that Rpt5 ubiquitination can be obstructed by Hsm3 in the assembled base and RP (Fig. 3I); the base and RP are dominant species in the isolated assembly intermediates (Fig. 4 A, ii, lane 13). These results are also attributed to sequential actions of Nas2 and Hsm3 during base assembly; Nas2 releases before completion of base, whereas Hsm3 remains in the assembled base (20). Indeed, Nas2 action during early-stage base assembly can be detected when NAS2 is deleted together with HSM3. The nas2Δhsm3Δ mutants exhibit an increased ratio of polyubiquitinated Rpt5 to unmodified Rpt5, compared with the hsm3Δ mutants (Fig. 4 A, i, compare lanes 9 and 11). This means that, in the nas2Δhsm3Δ mutants, Rpt5 ubiquitination is increased and Rpt5 incorporation into the base is decreased, relative to the hsm3Δ mutants. Our data demonstrate that Not4 ubiquitinates Rpt5 at two sequential stages of base assembly, upon vacancy of Nas2 in the early stage and Hsm3 in the late stage.
Ubiquitinated Rpt5 Antagonizes Incorporation of Ubp6 and Rpn1 into the Base.
Although ubiquitination typically serves as a rapid degradation signal for the conjugated protein (1), assembly intermediates harboring ubiquitinated Rpt5 are readily detectable (Fig. 4 A, i), indicating that these complexes are relatively stable and are not immediately targeted for degradation. Thus, we examined whether Not4-mediated ubiquitination of Rpt5 instead serves to regulate interactions between Rpt5 and the incoming subunits, thereby blocking the progression of base assembly (Fig. 1). To test this possibility, we tracked Rpn1, a non-ATPase subunit, which incorporates proximally to Rpt5 at the last step of base assembly but lacks its own chaperone (11–13, 30, 31). Rpn1 is a dual receptor for Ubp6 deubiquitinase and ubiquitinated protein substrates (32, 33), suggesting that regulation of Rpn1’s assembly is crucial for functional proteasome holoenzyme formation.
To track Rpn1 during base assembly, we used native PAGE to examine Rpn1-containing complexes in assembly intermediates from Fig. 4 A, i. Compared with wild type, the hsm3Δnas2Δrpn14Δ triple mutants exhibited substantially less Rpn1 in the base and RP, reflecting deficient base assembly (Fig. 4 A, ii, lane 1 vs. lane 3). However, when this mutant was combined with the not4-L35A allele, additional Rpn1-containing base precursors (BP*) were detected (Fig. 4 A, ii, lanes 3 and 4). Also, compared with the nas2Δ single mutant alone, the nas2Δnot4-L35A double mutant exhibited the Rpn1-containing BP* species at a greater level (Fig. 4 A, ii, lanes 13 and 14). This trend was also observed in the nas2Δrpn14Δ and nas2Δhsm3Δ double-deletion mutants (Fig. 4 A, ii, lanes 7–10). Together, these results support that Not4 competes with Nas2 for access to Rpt5 (Fig. 3 C and H) and that the resulting Rpt5 ubiquitination may hinder Rpn1 incorporation, thereby blocking the progression of base assembly (Fig. 1).
Because Rpn1 is a receptor for Ubp6 in the base (31–34), we examined whether Ubp6 association with assembly intermediates is also affected in Not4- vs. Not4-L35A–expressing cells in our panel of chaperone-deletion mutants. When Rpt5 is ubiquitinated, Ubp6 association is noticeably decreased in general (Fig. 4 A, iii, lanes 3, 7, 9, and 11). When Rpt5 is not ubiquitinated and Rpn1-containing species are detectable, Ubp6 association was restored in the corresponding assembly intermediates (Fig. 4 A, iii, lanes 4, 8, 10, and 12). Total protein levels of both Ubp6 and Rpn1 remain comparable in whole-cell lysates, indicating that their stability is not affected by Not4 (SI Appendix, Fig. S10). These results suggest that ubiquitinated Rpt5 may interfere with two sequential assembly events, incorporation of Rpn1 and then its ligand, Ubp6, during base assembly (SI Appendix, Fig. S11).
If ubiquitination of Rpt5 is responsible for blocking incorporation of Rpn1 and Ubp6 into the base, disabling Rpt5 ubiquitination should allow their incorporation, recapitulating the effect of the not4-L35A mutants in Fig. 4A. To test this, we substituted all three ubiquitination sites in Rpt5, K411, K426, and K428, with alanines; this mutant is referred to as rpt5-KA (Fig. 4B). In the rpt5-KA mutant alone, the Rpn1-containing BP* was detectable, as in the not4-L35A mutant (Fig. 4 B, ii, lanes 2 and 3), supporting that Not4-driven Rpt5 ubiquitination normally occurs during chaperone-mediated base assembly. In the hsm3Δnas2Δrpn14Δ cells, the rpt5-KA mutant abolished Rpt5 ubiquitination, indicating that these lysine residues are crucial for Not4-mediated ubiquitination of Rpt5 in vivo (Fig. 4 B, i, lanes 5 and 6). Importantly, in the hsm3Δnas2Δrpn14Δ cells, this rpt5-KA mutant led to restoration of both Rpn1 and Ubp6 in assembly intermediates, like the not4-L35A mutant (Fig. 4 B, ii and iii, lanes 5, 6, and 7). This finding supports that Rpt5 ubiquitination is indeed responsible for blocking both Rpn1 and Ubp6 incorporation during base assembly. In the nas2Δ background, the rpt5-KA mutant also exhibited Rpn1-containing BP*, like the not4-L35A mutant (Fig. 4 B, ii, lanes 10 and 11). Thus, upon vacancy of Nas2 alone, Rpt5 ubiquitination can block Rpn1 incorporation, albeit transiently, because the other three chaperones (Rpn14, Hsm3, and Nas6) can still promote overall progression of base assembly (Fig. 4 B, ii, lanes 9–12; see base, RP). The resulting base and RP recruit Ubp6 substantially as they exist at a greater level than the Rpn1-BP* complex in the nas2Δ background. Nevertheless, relative to the nas2Δ mutant alone, a slight increase in Ubp6 incorporation can be detected in the nas2Δ rpt5-KA mutant, as in the nas2Δ not4-L35A mutant (Fig. 4 B, iii, lane 9 vs. lanes 10 and 11). These results support that Rpt5 ubiquitination is responsible for hindering Rpn1 and Ubp6 incorporation at the Nas2-dependent stage first and then further at the Hsm3-dependent stage, thereby blocking progression of base assembly events.
Not4-Driven Control on Ubp6 Assembly into the Proteasome Influences the Free Ubiquitin Pool.
Our data demonstrate that Not4 antagonizes incorporation of Rpn1 and Ubp6 during chaperone-mediated base assembly (Fig. 4 A and B). In the fully formed proteasome holoenzyme, Ubp6 regenerates ubiquitin from cellular ubiquitinated protein substrates, thereby sparing the ubiquitin from degradation by the proteolytic CP (32, 33, 35). Due to this function, Ubp6 deficiency results in perturbation in the global free ubiquitin pool (32, 36). We examined whether Not4-mediated control on Ubp6 during proteasome assembly can be reflected in the free ubiquitin level of the cell. We assessed both free ubiquitin and ubiquitin conjugates using whole-cell lysates from a panel of chaperone-deletion mutants harboring Not4 or Not4-L35A. In all chaperone mutants, polyubiquitin conjugates generally accumulated due to dysfunctional proteolysis resulting from deficient proteasome assembly (Fig. 4C, lanes 3–12) (11–13). However, free ubiquitin is decreased in each chaperone mutant and is increased in its corresponding not4-L35A mutant (Fig. 4C, lanes 3–12; see quantification in the graph). This trend agrees well with reduced Ubp6 incorporation during base assembly in the chaperone mutants and its restoration in their corresponding Not4-L35A–expressing cells (Fig. 4 A, iii; also see SI Appendix, Fig. S12). These data demonstrate that Not4-mediated control of Ubp6 incorporation into the proteasome is also reflected in Ubp6’s activity in the proteasome in maintaining the global free ubiquitin pool.
Discussion
It is well established that four evolutionarily conserved chaperones (Nas2, Hsm3, Rpn14, and Nas6) orchestrate maturation of the proteasome holoenzyme by promoting assembly of the nine-subunit base (12–14, 23). The chaperones bind to specific Rpt proteins and facilitate heterohexameric Rpt ring assembly of the base, while blocking its premature association with the lid and CP (10, 11, 15–18). Here, we show that chaperone-dependent base assembly utilizes a specific ubiquitination-mediated control through the Not4 ubiquitin ligase.
Our findings demonstrate that Not4 provides a surveillance mechanism for proper assembly of the heterohexameric Rpt ring by monitoring Not4’s ubiquitination target, Rpt5. Not4-specific ubiquitination sites in Rpt5 are obstructed sequentially, first by Nas2, and then by Hsm3 (Figs. 3 and 4). Therefore, Not4 can selectively detect assembly intermediates that proceed without the chaperones in a stepwise manner during base assembly (Fig. 4 and SI Appendix, Fig. S13). Although Nas6 and Rpn14 do not directly obstruct Rpt5 ubiquitination sites (Fig. 3 A and D), they can limit accumulation of Rpt5-containing intermediates by promoting overall progression of assembly events into the proteasome holoenzyme (Fig. 1A) (10–13, 17). Thus, any excess Rpt5-containing complexes reflect not only any deficiency of chaperones but also untimely assembly events; both can be recognized by Not4. The importance of this mechanism is supported by features of chaperone-mediated proteasome assembly. In the cell, the chaperones are present in substoichiometric levels to proteasome subunits (37). Availability of chaperones is controlled mainly by recycling; they release upon completion of proteasome holoenzyme assembly for a new round of assembly (10, 11, 16, 17). Moreover, multiple chaperones act cooperatively to maintain a high rate of proteasome assembly and activity (11–14, 16, 23). Our findings suggest that Not4 may monitor each round of proteasome assembly and provide a checkpoint for any potentially defective assembly intermediates until the chaperones can act properly on them.
Not4-mediated ubiquitination of Rpt5 antagonizes the progression of base assembly by blocking incorporation of two crucial subunits, Rpn1 and Ubp6, which together process ubiquitinated protein substrates in the proteasome (Fig. 4) (32, 33, 35). Given that Rpt5 and Rpn1 are closely positioned in the base (30, 31), ubiquitinated Rpt5 might lower Rpn1 affinity to the base. In line with a receptor–ligand relationship of Rpn1-Ubp6, Rpt5 ubiquitination also controls Ubp6 binding to the base (Fig. 4 and SI Appendix, Figs. S12 and S13). Both positioning and activity of Ubp6 are affected by multiple conformational changes of the heterohexameric Rpt ring during the ring’s assembly via the chaperones (17, 28, 31, 34). Our data reveal a regulatory point during base assembly in that Not4 ensures the accurate assembly of the heterohexameric Rpt ring, before its progression to the higher-order complexes. It remains to be determined how arrested assembly intermediates might be dealt with, especially if the arrested states are prolonged in the cell. Cellular structures, referred to as foci or granules, have been shown to store excess proteasomes and their subcomplexes, such as RP and CP, and also to form during autophagy-mediated degradation of these complexes (38–41). We speculate that cells might additionally sequester the arrested assembly intermediates in such foci or granules, where these complexes could further await opportunities for rectification of the assembly intermediates’ defects or, alternatively, undergo degradation in the event that these defects cannot be repaired.
Materials and Methods
A complete list of yeast strains is provided in SI Appendix, Table S3. Detailed procedures for ubiquitination assays and affinity purification of assembly intermediates are described in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jeroen Roelofs, Dan Finley, Keiji Tanaka, Yasushi Saeki, Martine Collart, and Anja-Katrin Bielinsky for sharing antibodies, plasmids, E. coli strains, and yeast strains. We also thank James Orth for providing helpful comments on the manuscript. This study was supported by Department of Defense/Defense Advanced Research Projects Agency (DARPA) Grant DARPA-13-34-RTA-FP-007 (to W.O.), the Boettcher Webb-Waring Biomedical Research Award (to S.P.), and NIH Grant 1R01GM127688-01A1 (to S.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805353115/-/DCSupplemental.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 3.Kish-Trier E, Hill CP. Structural biology of the proteasome. Annu Rev Biophys. 2013;42:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lasker K, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci USA. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 7.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 8.Smith DM, et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park S, et al. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roelofs J, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saeki Y, Toh-E A, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneko T, et al. Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Barrault MB, et al. Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proc Natl Acad Sci USA. 2012;109:E1001–E1010. doi: 10.1073/pnas.1116538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S, et al. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature. 2013;497:512–516. doi: 10.1038/nature12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F, et al. Nucleotide-dependent switch in proteasome assembly mediated by the Nas6 chaperone. Proc Natl Acad Sci USA. 2017;114:1548–1553. doi: 10.1073/pnas.1612922114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokolova V, Li F, Polovin G, Park S. Proteasome activation is mediated via a functional switch of the Rpt6 C-terminal tail following chaperone-dependent assembly. Sci Rep. 2015;5:14909. doi: 10.1038/srep14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Förster F, et al. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun. 2009;388:228–233. doi: 10.1016/j.bbrc.2009.07.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: Implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satoh T, et al. Structural basis for proteasome formation controlled by an assembly chaperone nas2. Structure. 2014;22:731–743. doi: 10.1016/j.str.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Hanssum A, et al. An inducible chaperone adapts proteasome assembly to stress. Mol Cell. 2014;55:566–577. doi: 10.1016/j.molcel.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousseau A, Bertolotti A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536:184–189. doi: 10.1038/nature18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakata E, et al. The catalytic activity of Ubp6 enhances maturation of the proteasomal regulatory particle. Mol Cell. 2011;42:637–649. doi: 10.1016/j.molcel.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 25.Panasenko OO, Collart MA. Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol Cell Biol. 2011;31:1610–1623. doi: 10.1128/MCB.01210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulder KW, et al. Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics. 2007;176:181–192. doi: 10.1534/genetics.106.060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SY, De la Mota-Peynado A, Roelofs J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. J Biol Chem. 2011;286:36641–36651. doi: 10.1074/jbc.M111.280875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, et al. Conformational landscape of the p28-bound human proteasome regulatory particle. Mol Cell. 2017;67:322–333.e6. doi: 10.1016/j.molcel.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besche HC, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33:1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweitzer A, et al. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc Natl Acad Sci USA. 2016;113:7816–7821. doi: 10.1073/pnas.1608050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bashore C, et al. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol. 2015;22:712–719. doi: 10.1038/nsmb.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y, et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016;351:aad9421. doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aufderheide A, et al. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci USA. 2015;112:8626–8631. doi: 10.1073/pnas.1510449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 36.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 38.Laporte D, Salin B, Daignan-Fornier B, Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol. 2008;181:737–745. doi: 10.1083/jcb.200711154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall RS, McLoughlin F, Vierstra RD. Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep. 2016;16:1717–1732. doi: 10.1016/j.celrep.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 40.Marshall RS, Vierstra RD. Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. eLife. 2018;7:e34532. doi: 10.7554/eLife.34532. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Waite KA, De-La Mota-Peynado A, Vontz G, Roelofs J. Starvation induces proteasome autophagy with different pathways for core and regulatory particles. J Biol Chem. 2016;291:3239–3253. doi: 10.1074/jbc.M115.699124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unverdorben P, et al. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci USA. 2014;111:5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Singh CR, et al. 1.15 Å resolution structure of the proteasome-assembly chaperone Nas2 PDZ domain. Acta Crystallogr F Struct Biol Commun. 2014;70:418–423. doi: 10.1107/S2053230X14003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.