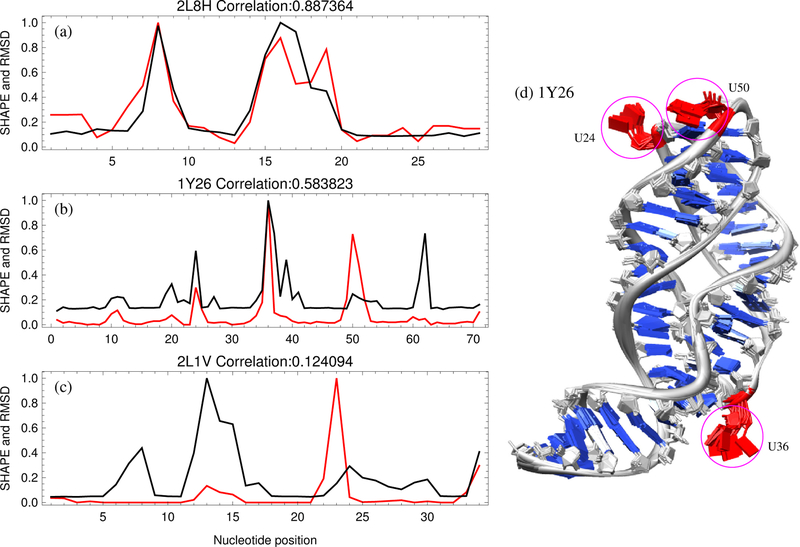
Figure 2:
Experimental SHAPE data is generally correlated with nucleotide dynamics. The scaled SHAPE reactivities30, 36, 37 (shown in red) as a function of nucleotide position compared with the MD-estimated nucleotide flexibilities (scaled RMSD shown in black) for the highest correlated case (a) 2L8H, the example case (b) 1Y26, and the lowest correlated case (c) 2L1V, respectively. (d) Ten snapshots of an MD simulation for 1Y26, with backbone phosphate (P) atoms position-constrained. The global fold of the RNA molecule is conserved, while each nucleotide has different fluctuations around its equilibrium state due to the constraints provided by other nucleotides. Highlighted in red are three nucleotides (U24, U36 and U50) that have large fluctuations and the highest SHAPE reactivity. The ligand molecules are not included during MD simulations.