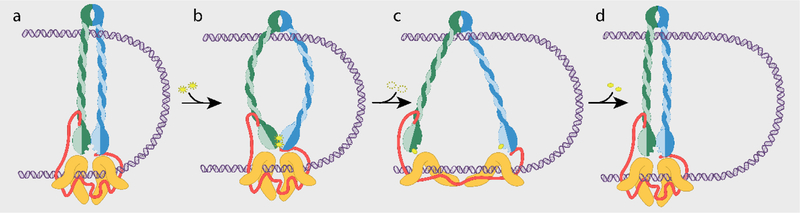
Figure 2: The Tethered Inchworm Model.
a, The SMC complex is composed of two SMC proteins (green and blue) which dimerize at the hinge (top) and at the head domain (bottom). Tethering the two heads together is a kleisin (red) further bound by HAWK proteins (orange). The SMC complex forms a small loop in the DNA (purple) by binding with both the hinge dimer and the kleisin-HAWK subcomplexes. b, Binding of 2 ATP molecules (yellow) by the ATPase head domains (green and blue) induces a conformational rotation of each head. This movement forces the coiled-coil arms apart, bending them, and propagating the strain to the hinge domains. c, ATP hydrolysis causes dissociation of the head domains and opening of the SMC arms. In this model, the leading HAWK would slide forward along the DNA due to its weaker affinity for DNA. The kleisin would straighten and unfurl to accommodate this movement and in doing so pull on the HAWKs, stretching these spring-like proteins. d, In the extended configuration the DNA binding affinities of the HAWKs then reverse causing the lagging HAWK to catch up as the head domains reunite, completing a mechanochemical cycle that has enlarged the DNA loop.