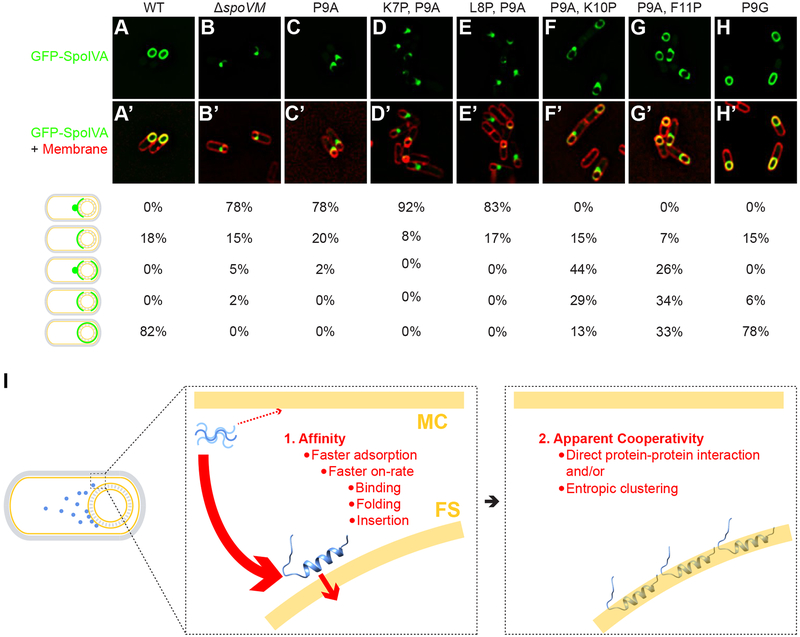
Figure 3: Proper localization of GFP-SpoIVA in vivo requires properly localized SpoVM.
Localization of GFP-SpoIVA in sporulating B. subtilis cells in the presence (A; strain KR165) or absence (B; KR178) of SpoVM, or in the presence of SpoVMP9A (C; SE50), SpoVMK7P,P9A (D; EYKS919), SpoVML8P,P9A (E; EYKS920), SpoVMP9A,K10P (F; EYKS892), SpoVMP9A,F11P (G; EYKS893), or SpoVMP9G (H; EYKS929). (A’-H’) Overlay of GFP fluorescence (green) and membranes (red) visualized with fluorescent dye FM4–64 from panels A-H, respectively. Fraction of cells displaying the indicated localization pattern is shown to the right (n > 55 cells for each). (I) Model for the affinity/cooperativity-driven localization of SpoVM onto positively curved membranes. Left: depiction of sporulating B. subtilis in which SpoVM molecules (blue) synthesized in the mother cell (MC) adsorb onto the convex membrane surface (yellow) of the forespore (FS). Center: unordered SpoVM molecule (blue, upper left) has a higher intrinsic affinity for the convex forespore surface (thick red arrow). Higher affinity is mediated by a faster on-rate, which is composed of three steps: initial binding, protein folding, and membrane insertion. We propose that folding and/or insertion occur more quickly on membranes of preferred curvature. Right: successful insertion of the first SpoVM molecule positively influences the subsequent insertion of other SpoVM molecules, mediated by direct protein-protein interaction and/or entropic clustering.