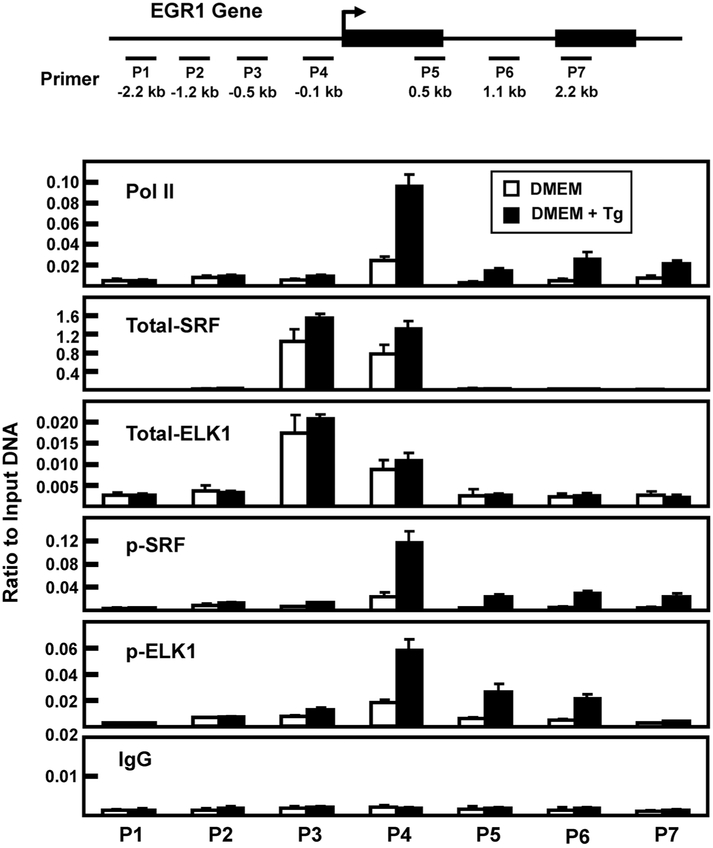
Fig. 6.
Association of RNA polymerase II, SRF, and ELK1 at the EGR1 promoter after 2 h of Tg treatment. The locations of primers (labeled P1-P7) used to analyze the human EGR1 gene are illustrated relative to the transcription start site (arrow) and the two exons that comprise the protein-coding region of the gene are shown as black rectangles. The primer sequences are listed in Table 1. HepG2 cells were incubated in DMEM (Control) or DMEM + Tg for 2 h and then the cells were subjected to ChIP analysis with antibodies specific for RNA Pol II, total SRF, p-SRF, total ELK1, p-ELK1, and a non-specific IgG as a negative control. The data are plotted as the ratio to input DNA and are the averages ± SD for at least three samples within an experiment. The data shown are representative of multiple independent experiments.