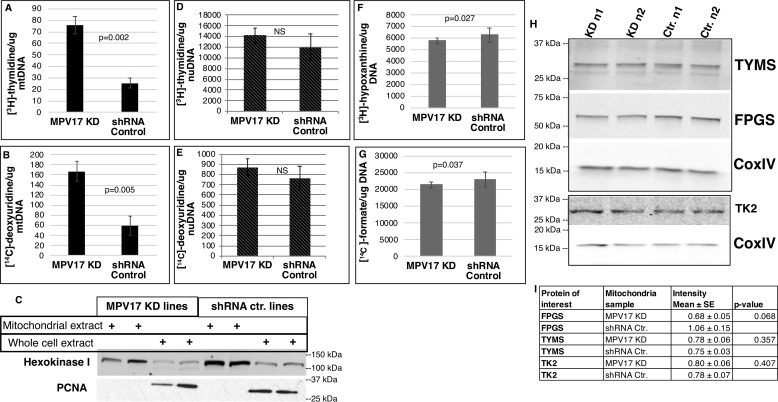
Figure 3.
MPV17 KD increased mitochondrial dTMP synthesis. A, [3H]thymidine incorporation into newly synthesized mtDNA via salvage pathway: KD, n = 3; control, n = 2. B, [14C]deoxyuridine incorporation into mtDNA via folate-dependent de novo dTMP pathway: KD, n = 3; control, n = 2. C, mtDNA was extracted from isolated mitochondria. Representative Western blotting of protein extracts of mitochondrial fractions and respective whole-cell extracts is shown. Mitochondrial fractions were free of nuclear contamination (proliferating cell nuclear antigen (PCNA) (nuclear marker) and hexokinase I (mitochondrial marker)). D and E, [3H]thymidine incorporation into newly synthesized nuDNA via salvage pathway (D) and [14C]deoxyuridine incorporation into nuDNA via folate-dependent de novo dTMP pathway (E): n = 4 each group, not significant (NS). F and G, newly synthesized nuDNA containing purines made via the salvage pathway (F) and the folate-dependent de novo synthesis pathway (G), as quantified by [3H]hypoxanthine and [14C]formate incorporation, respectively; n = 6 each group. A, B, and D–G, 3H and 14C channels were counted in dual DPM mode on a scintillation counter. H and I, Western blots of mitochondrial protein extracts from different biological replicates probed for TK2, TYMS, and FPGS. Densitometry was performed using ImageJ. The intensities of nonsaturated bands were quantified and normalized to CoxIV, which served as a mitochondrial marker and mitochondrial protein-loading control. Data are shown as mean ± S.D. (error bars) (A–H) or means ± S.E. (error bars) (I). Statistical significance was determined by a two-tailed Student's t test (NS, p > 0.05). Ctr., scrambled shRNA control.