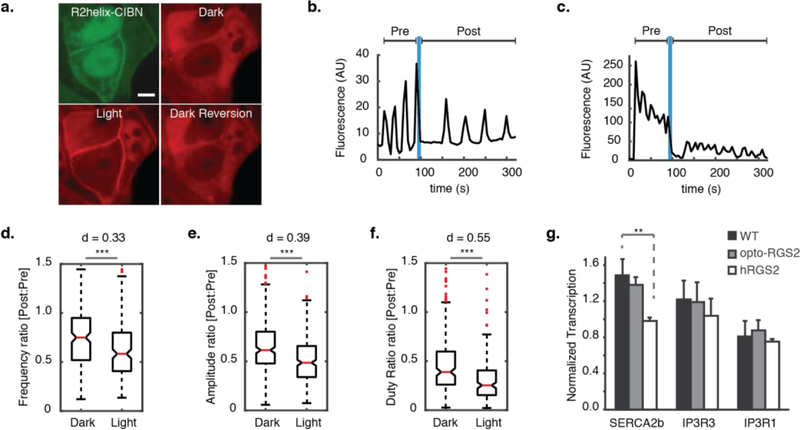
Figure 3. Inhibitory regulation of calcium oscillations in opto-RGS2 cell lines.
(a) Fluorescence micrographs of light-induced translocation of mCherry-tagged CRY2-R2box (red) in opto-RGS2 cell lines. (top left) GFP-tagged R2helix-CIBN is membrane- and nuclear-localized, consistent with known R2helix distribution patterns. (top right) CRY2-R2box in the cytoplasm of the same cell, (bottom left) membrane localization 30 seconds after brief (1 s) blue light stimulation, and (bottom right) thermal reversion back to the cytoplasm in the dark (900 s post-illumination). Scale = 5 um. (b) Example X-rhodamine calcium imaging traces of dynamic suppression of 100 uM CCh-induced oscillations in response to a single epoch of blue light. Optogenetic dampening primarily decreased the frequency. (c) Same as panel b, except where optogenetic dampening primarily decreased the amplitude. (d) Single epoch of blue light dynamically reduced oscillation frequency. Post-illumination parameter values were normalized to pre-illumination values of the same oscillation in the same cell, as in panel a. (*** p < 0.001, d = Cohen’s effect size) (e) Same as panel d, except for amplitude. (f) Same as panel d, except for duty ratio. (g) Quantitative PCR measurements of SERCA2b and IP3R transcripts. SERCA2b level was altered in hRGS2 cells to compensate for long-term reduction in calcium load (p < 0.01), but not in opto-RGS2 cells.