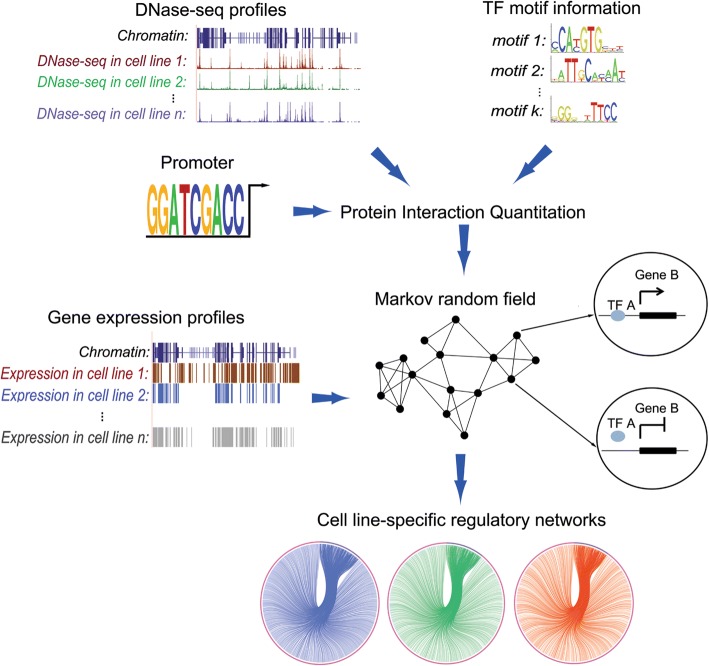
Fig. 1.
Workflow of the construction of 110 cell line-specific regulatory networks. Firstly, perform PIQ on DNase-seq profiles and TF motifs to predict genome-wide transcription factor binding sites for 368 TFs in 110 cell lines, respectively. Secondly, map these transcription factor binding sites to the promoter regions of genes and thus link TFs to target genes. Thirdly, construct regulatory networks with a Markov random field (MRF) model based on the cell line similarity measured by the expression profiles of these cell lines. Finally, we get the cell line-specific regulatory network for each cell line