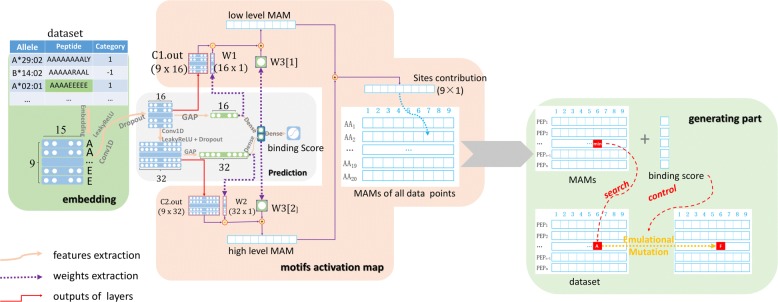
Fig. 1.
Pipeline of our Motifs Activation Map network. Embedding step is to encode each amino acid into a 15-dimension vector. Prediction step is to predict the binding probability from 0 (non-binder) to 1 (binder) and the weights of MAM network. Our MAM network is to calculate the contribution scores at each site then generate new peptides with mutating the amino acid with lowest contribution score. Here we take 9-mer as an example