Abstract
Background
Alfalfa (Medicago sativa L.) is a forage legume with significant agricultural value worldwide. MicroRNAs (miRNAs) are key components of post-transcriptional gene regulation and essentially regulate many aspects of plant growth and development. Although miRNAs were reported in alfalfa, their expression profiles in different tissues and the discovery of novel miRNAs as well as their targets have not been described in this plant species.
Results
To identify tissue-specific miRNA profiles in whole plants, shoots and roots of three different alfalfa genotypes (Altet-4, NECS-141and NF08ALF06) were used. Small RNA libraries were generated and sequenced using a high-throughput sequencing platform. Analysis of these libraries enabled identification of100 miRNA families; 21 of them belong to the highly conserved families while the remaining 79 families are conserved at the minimum between M. sativa and the model legume and close relative, M. truncatula. The profiles of the six abundantly expressed miRNA families (miR156, miR159, miR166, miR319, miR396 and miR398) were relatively similar between the whole plants, roots and shoots of these three alfalfa genotypes. In contrast, robust differences between shoots and roots for miR160 and miR408 levels were evident, and their expression was more abundant in the shoots. Additionally, 17 novel miRNAs were identified and the relative abundance of some of these differed between tissue types. Further, the generation and analysis of degradome libraries from the three alfalfa genotypes enabled confirmation of 69 genes as targets for 31 miRNA families in alfalfa.
Conclusions
The miRNA profiles revealed both similarities and differences in the expression profiles between tissues within a genotype as well as between the genotypes. Among the highly conserved miRNA families, miR166 was the most abundantly expressed in almost all tissues from the three genotypes. The identification of conserved and novel miRNAs as well as their targets in different tissues of multiple genotypes increased our understanding of miRNA-mediated gene regulation in alfalfa and could provide valuable insights for practical research and plant improvement applications in alfalfa and related legume species.
Introduction
Alfalfa (Medicago sativa L.) is an important forage legume species with global adaptation, high forage quality and the capacity for harvesting biomass multiple times during the growing season. Alfalfa is an autotetraploid (2n = 4x = 32), perennial outcrossing species with high levels of genetic diversity in cultivated and non-cultivated populations. Besides its use as a forage, alfalfa also has potential crop for biofuel production [1]. Alfalfa has the capacity for symbiotic nitrogen fixation and can also contribute to reduce soil erosion [2, 3].
Endogenous non-coding RNAs of approximately 21–22 nucleotides represent plant miRNAs that silence gene expression by binding to complementary sequences of its target mRNA at the post-transcriptional level. Such targeting results in mRNA cleavage and degradation or repression of translation, with the former being more prevalent in plants [4–7]. The miRNA analyses in different plant species highlight the important regulatory roles of miRNAs in multiple organs (roots, stems, leaves and flowers), differentiation and development, leaf polarity, transition from juvenile to vegetative stages and vegetative to reproductive phases, and regulation of plant responses to biotic and abiotic stresses [8–10].
Several investigations have shown that plant miRNAs can be classified into conserved and novel lineage- or species-specific miRNAs. Conserved miRNAs and their corresponding target genes are commonly found in all or most angiosperms, with some also being described in gymnosperms as well as primitive land plants such as ferns [11, 12]. However, miRNA analysis in several legumes including M. truncatula, soybean (Glycine max L), chickpea (Cicer arietinum L.), common bean (Phaseolus vulgaris), and Lotus japonicus indicate the presence of miRNAs that seem to be specific to certain legumes and there could have important gene regulatory roles [13–19]. Although recent attempts were made to report miRNAs from alfalfa (M. sativa) [20–22], these do not include the discovery of novel miRNAs, and most importantly, the miRNA target genes have not been confirmed in this legume species. Understanding miRNAs and their target gene regulation in various tissues can provide further insights into the miRNA target networks operating in a tissue-specific manner in alfalfa.
In order to identify conserved miRNAs as well as novel miRNAs from alfalfa, we constructed and sequenced small RNA libraries from whole clonally propagated plants, roots and shoots of three alfalfa genotypes (Altet-4, NECS-141 and NF08ALF06). The sequenced reads were mapped to known miRNAs in M. truncatula, deposited in the miRBase to identify and annotate the miRNAs in alfalfa. Degradome libraries were constructed and sequenced from these three genotypes to characterize the miRNA gene targets.
Materials and methods
Plant materials and growth conditions
Three alfalfa genotypes NECS-141, Altet-4 and NF08ALF06 were evaluated in this study. NECS-141 is the genotype being used to sequence the tetraploid alfalfa genome [23]. Altet-4 is an aluminum tolerant genotype used to develop a mapping population [24]. NF08ALF06 is a commercial breeding line with good agronomic performance (Forage Genetics International). The three alfalfa genotypes (NECS-141, Altet-4 and NF08ALF06) were clonally propagated and grown in tissue culture. After 13 d of growth in rooting media, these were transferred to medium at pH 7 for 96 h as previously described [25]. The rooting media contains 0.55 g/L Murashige & Skoog Basal Medium with Vitamins (PhytoTechnology #M519), 1 ml Plant Preservative Mixture, PPM (PhytoTechnology), adjust the pH to 5.8, and add 12 g/L Gelzan. The plants were placed in a Conviron growth chamber (24 °C, 18 h /6 h day/night cycle, 100 μmol light intensity) for root development and growth. An additional 20 clonally propagated plants of these genotypes were grown in a Conviron growth chamber as previously described and used to evaluate the tissue-specific expression of the miRNAs. Tissue samples were harvested and immediately flash frozen in liquid nitrogen and stored at − 80 °C.
Small RNA library construction and sequencing
Total RNA was isolated from the whole plants, roots and shoots of three alfalfa genotypes using TRIzol ® Reagent (Invitrogen), according to the manufacturer’s instructions. The quality of total RNA was monitored on 1% agarose gel and their concentrations were measured using Nanodrop spectrophotometer. Small RNA libraries were generated as described previously [26] by following the protocol described for the Illumina Truseq® Small RNA Preparation kit (Illumina, San Diego, USA). Briefly, 1 μg total RNA per sample was ligated sequentially with 5′ and 3’ RNA adaptors. The ligated products were converted into cDNAs and then amplified using PCR. The amplified products were sequenced using an Illumina Hiseq® Analyzer.
Identification of conserved and novel miRNAs
The raw sequencing reads were processed as follows: adaptor sequences were trimmed off from the raw reads to obtain small RNAs. These reads were then mapped to ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNAs (snRNA), and the aligned and mapped reads were not used for further analysis. The remaining reads were aligned to miRBase v 20 [27] to identify miRNAs in M. sativa. The reads with 100% sequence identity were designated as conserved miRNA homologs. To identify novel miRNAs, the presence of the miRNA-star (miRNA*) sequences coupled with the predictable hairpin-like structure for the precursor sequences were used.
Degradome library construction and analyses
Degradome libraries from the alfalfa genotypes NECS-141, Altet-4 and NF08ALF06 were constructed as previously described to identify potential target mRNAs [28]. Briefly, the cleaved 5′ monophosphate containing polyadenylated mRNA fragments were ligated to an RNA oligo-nucleotide adapter containing MmeI recognition site at its 3′ end. The ligated products were converted into cDNA using reverse transcriptase and the product was amplified using only 5 PCR cycles. The PCR product was eluted, digested with MmeI restriction enzyme and then ligated to a double-stranded DNA adapter. The ligated product was again purified and amplified using 15 cycles of PCR. The final PCR product was sequenced. The reads were processed for quality and then aligned to the transcriptome assembly of M. truncatula to identify potential miRNA targets using the SeqTar pipeline [29].
Results and discussion
The analyses of small RNA libraries
High-throughput sequencing has been used to identify miRNAs and their target mRNAs in plants [15, 30, 31]. To catalogue conserved and novel miRNAs in alfalfa, a total of eight small RNA libraries from the whole plants, roots and shoots of Altet-4, NECS-141 and NF08ALF06 genotypes were constructed and sequenced. After removal of the adapter sequences and low-quality reads, the total reads ranging between 11 to 42 million, and unique reads ranging between 1.8 to 8.5 million reads from these nine libraries were obtained (Table 1). However, the quality of the small RNA library generated from the shoots of NF08ALF06 did not meet the threshold criteria, therefore only NECS-141 and Altet-4 were used for the miRNA analyses of shoot tissues.
Table 1.
The mapping of total and unique reads obtained from different small RNA libraries
| Altet-4 whole plants | NECS-141 whole plants | NF08ALF06 whole plants | Altet-4 Roots | NECS-141 Roots | NF08ALF06 Roots | Altet-4 Shoots | NCES-141 Shoots | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total reads | Unique reads | Total reads | Unique reads | Total reads | Unique reads | Total reads | Unique reads | Total reads | Unique reads | Total reads | Unique reads | Total reads | Unique reads | Total reads | Unique reads | |
| cdna | 6,858,719 | 336,266 | 6,197,308 | 430,819 | 8,142,985 | 493,929 | 9,352,477 | 276,276 | 19,665,339 | 732,738 | 8,930,063 | 352,167 | 17,117,689 | 909,491 | 10,943,411 | 669,572 |
| ncRNAs | 6,810,937 | 261,134 | 5,666,067 | 213,396 | 7,722,727 | 284,785 | 9,633,993 | 271,496 | 18,454,661 | 272,596 | 8,834,028 | 237,827 | 14,718,199 | 289,087 | 7,855,629 | 128,514 |
| pre-miRBase | 567,518 | 3182 | 943,976 | 3888 | 1,162,233 | 4005 | 147,326 | 2409 | 1,102,879 | 5780 | 426,327 | 3213 | 2,520,492 | 7051 | 3,840,771 | 6449 |
| repeats | 5,451,840 | 162,552 | 4,218,992 | 148,297 | 5,756,349 | 183,418 | 8,267,063 | 158,310 | 15,744,403 | 180,030 | 7,536,403 | 146,043 | 10,855,687 | 192,377 | 3,798,312 | 104,708 |
| genome | 8,951,430 | 1,142,594 | 9,878,838 | 2,398,705 | 11,387,413 | 2,053,582 | 11,557,742 | 784,140 | 29,143,549 | 5,078,322 | 11,588,832 | 1,488,546 | 28,744,231 | 5,246,027 | 29,192,098 | 5,834,731 |
| total | 12,008,892 | 2,343,120 | 11,645,217 | 3,348,188 | 15,733,102 | 3,739,163 | 14,377,336 | 1,860,736 | 33,335,201 | 6,947,622 | 14,378,859 | 2,708,737 | 42,196,888 | 8,564,218 | 34,441,313 | 7,748,996 |
Quantification of miRNA abundances between the genotypes and tissues was preceded by normalizing the expression levels of miRNA families to reads per ten million (RPTM). The normalized miRNA family read frequencies ranged between 1 to 552,267 RPTM for the whole plants, between 1 to 134,679 RPTM for the root samples, and 1 to 165,310 RPTM for the shoot samples (Table 2). The range of miRNA read frequencies varied slightly between the three genotypes. As expected, the most conserved miRNAs appeared to be the most abundantly expressed in all tissues and genotypes, with the exception of miR169, miR393, miR395 and miR172 which exhibited low abundances. Specifically, miR172 levels in roots and shoots of the three genotypes were extremely low and in most cases was below 20 RPTM (Table 2). The miRNA families with the lowest expression levels, and in some cases as low as 1 RPTM, were largely represented by the non-conserved miRNAs or miRNAs that have been reported exclusively from M. truncatula (miRBase) that include miR2601, miR2674, miR5207, miR5241, miR5243, miR5244, miR5255, miR5257, miR5269, miR5282, miR5289, miR5294, miR5296, miR5299, miR5561, miR5744, and miR7701 (Table 2). miR5207 is the only miRNA that was also reported from Gossypium raimondii (miRBase). The majority of the miRNA families identified are 21 nt long, although some cases including miR2601 and miR2603 were represented by 22 nucleotides. Further, a total of 23 miRNA families included between miR5267 to miR5299 were 24 nt long. The fact that these small RNAs were initially identified in M. truncatula (miRBase), and could be identified in several independent small RNA libraries from three different alfalfa genotypes (Table 2), suggests that these sequences and their associated processing are conserved between alfalfa and its close relative M. truncatula. However, their extremely low abundances coupled with their longer read lengths could also indicate that these may be 24-nt long siRNAs. Additional studies are needed to assess the precise nature of these small RNAs, i.e., miRNAs or siRNAs.
Table 2.
Identified miRNA families and their frequencies (reads per ten million [RPTM]) in whole plants, roots and shoots of three alfalfa genotypes (miRNA-stars were marked in bold)
| Whole plants | Roots | Shoots | ||||||
|---|---|---|---|---|---|---|---|---|
| Altet-4 | NECS-141 | NF08ALF06 | Altet-4 | NECS-141 | NF08ALF06 | Altet-4 | NECS-141 | |
| miR156-5p | 4712 | 7243 | 6436 | 1001 | 3466 | 3145 | 19,808 | 47,306 |
| miR156-3p | 3262 | 4012 | 2992 | 545 | 755 | 548 | 4634 | 6420 |
| miR159-3p | 6315 | 11,050 | 8484 | 3910 | 23,465 | 10,549 | 61,929 | 103,370 |
| miR160-5p | 225 | 417 | 351 | 20 | 277 | 113 | 3505 | 8706 |
| miR162-3p | 140 | 229 | 292 | 194 | 454 | 361 | 533 | 517 |
| miR164-5p | 108 | 275 | 306 | 6 | 77 | 57 | 48 | 431 |
| miR166-3p | 336,905 | 552,267 | 534,054 | 34,634 | 111,596 | 134,679 | 101,118 | 131,196 |
| miR166-5p | 544 | 960 | 614 | 228 | 508 | 438 | 800 | 1216 |
| miR167-5p | 218 | 470 | 722 | 107 | 240 | 357 | 699 | 1389 |
| miR167-3p | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| miR168-5p | 1121 | 1980 | 1691 | 735 | 2960 | 1317 | 3460 | 5049 |
| miR168-3p | 672 | 691 | 768 | 182 | 443 | 194 | 5550 | 5638 |
| miR169-5p | 19 | 34 | 35 | 47 | 55 | 35 | 46 | 59 |
| miR169-3p | 7 | 12 | 5 | 6 | 18 | 7 | 2 | 2 |
| miR171-3p | 51 | 120 | 232 | 44 | 238 | 316 | 60 | 85 |
| miR171e-5p | 26 | 39 | 44 | 22 | 37 | 42 | 7 | 6 |
| miR172-3p | 62 | 138 | 240 | 0 | 1 | 1 | 2 | 3 |
| miR172-5p | 3 | 8 | 20 | 1 | 1 | 2 | 2 | 2 |
| miR319-3p | 1631 | 3689 | 2101 | 1607 | 6281 | 3323 | 4330 | 10,864 |
| miR319-5p | 46 | 72 | 74 | 3 | 20 | 14 | 129 | 559 |
| miR390-5p | 95 | 410 | 318 | 86 | 656 | 234 | 121 | 382 |
| miR393-5p | 11 | 24 | 34 | 4 | 8 | 10 | 22 | 45 |
| miR395-3p | 3 | 8 | 7 | 12 | 13 | 7 | 2 | 0 |
| miR396-5p | 12,185 | 21,926 | 22,411 | 2835 | 14,549 | 8121 | 39,236 | 58,336 |
| miR396-3p | 250 | 437 | 437 | 76 | 312 | 188 | 323 | 356 |
| miR397-5p | 57 | 28 | 15 | 37 | 16 | 11 | 94 | 61 |
| miR398a-5p | 19 | 16 | 25 | 0 | 2 | 1 | 4 | 3 |
| miR398-3p | 3814 | 3223 | 2272 | 2101 | 4086 | 3176 | 35,538 | 26,478 |
| miR399-3p | 17 | 11 | 11 | 25 | 26 | 13 | 62 | 43 |
| miR408-3p | 2656 | 1301 | 1096 | 977 | 737 | 570 | 6380 | 2866 |
| miR408-5p | 17 | 7 | 12 | 12 | 14 | 8 | 55 | 35 |
| miR482-3p | 28 | 27 | 49 | 18 | 19 | 45 | 41 | 105 |
| miR482-5p | 7 | 10 | 10 | 11 | 19 | 13 | 9 | 12 |
| miR530-5p | 2 | 7 | 8 | 0 | 1 | 1 | 2 | 4 |
| miR1507–3 | 963 | 1789 | 1701 | 881 | 1596 | 1230 | 1778 | 3349 |
| miR1510-5p | 1959 | 4278 | 3520 | 523 | 3505 | 1429 | 12,496 | 34,705 |
| miR1510-3p | 96 | 151 | 167 | 52 | 118 | 63 | 256 | 617 |
| miR2111 | 47 | 20 | 10 | 44 | 15 | 42 | 278 | 22 |
| miR2118 | 5607 | 11,948 | 16,134 | 106 | 610 | 307 | 79,977 | 165,310 |
| miR2199 | 95 | 15 | 42 | 21 | 18 | 30 | 156 | 13 |
| miR2585 | 57 | 7 | 74 | 28 | 1 | 22 | 239 | 10 |
| miR2587 | 0 | 6 | 9 | 0 | 10 | 10 | 13 | 28 |
| miR2590 | 15 | 41 | 42 | 23 | 55 | 25 | 109 | 177 |
| miR2592 | 393 | 1350 | 395 | 119 | 1612 | 268 | 1224 | 1742 |
| miR2601-5p | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| miR2603-5p | 0 | 8 | 1 | 1 | 1 | 1 | 5 | 24 |
| miR2629-5p | 2 | 5 | 4 | 1 | 3 | 7 | 2 | 5 |
| miR2632-5p | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 18 |
| miR2634-3p | 5 | 3 | 7 | 6 | 4 | 15 | 9 | 6 |
| miR2643-3p | 1502 | 2689 | 2106 | 382 | 1462 | 948 | 9682 | 24,971 |
| miR2651-3p | 27 | 52 | 22 | 4 | 21 | 7 | 40 | 49 |
| miR2661-5p | 3 | 4 | 5 | 2 | 4 | 5 | 13 | 9 |
| miR2666-3p | 0 | 21 | 0 | 0 | 14 | 0 | 0 | 29 |
| miR2674-3p | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| miR2678-3p | 2 | 6 | 4 | 0 | 4 | 4 | 4 | 12 |
| miR4414-3p | 2 | 4 | 4 | 0 | 1 | 1 | 3 | 7 |
| miR4414-5p | 1 | 3 | 4 | 1 | 1 | 0 | 5 | 7 |
| miR5037-5p | 4 | 3 | 13 | 3 | 8 | 24 | 2 | 4 |
| miR5204-3p | 4 | 10 | 6 | 3 | 28 | 17 | 6 | 10 |
| miR5205-5p | 7 | 22 | 14 | 0 | 6 | 6 | 15 | 6 |
| miR5207-5p | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| miR5208-3p | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 1 |
| miR5208d-5p | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| miR5211-5p | 432 | 85 | 23 | 559 | 71 | 41 | 292 | 59 |
| miR5213-5p | 801 | 836 | 887 | 181 | 891 | 829 | 1397 | 1379 |
| miR5214-3p | 63 | 155 | 153 | 97 | 414 | 452 | 153 | 201 |
| miR5225-5p | 4 | 2 | 8 | 3 | 1 | 8 | 1 | 1 |
| miR5230-5p | 1 | 2 | 1 | 0 | 1 | 0 | 6 | 1 |
| miR5231-5p | 10 | 7 | 7 | 3 | 11 | 1 | 43 | 69 |
| miR5232-5p | 67 | 253 | 419 | 56 | 503 | 417 | 602 | 3964 |
| miR5237-3p | 2 | 2 | 0 | 0 | 2 | 1 | 6 | 4 |
| miR5238-5p | 2 | 0 | 2 | 1 | 2 | 1 | 0 | 0 |
| miR5239-5p | 347 | 269 | 430 | 16 | 52 | 72 | 622 | 773 |
| miR5241-3p | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| miR5243-3p | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| miR5244-3p | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| miR5248-5p | 0 | 2 | 1 | 0 | 2 | 1 | 0 | 3 |
| miR5255-3p | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| miR5257-5p | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| miR5261-3p | 76 | 89 | 93 | 22 | 302 | 127 | 283 | 227 |
| miR5266-5p | 0 | 0 | 0 | 4 | 2 | 3 | 0 | 1 |
| miR5267-5p | 1 | 3 | 1 | 0 | 1 | 1 | 0 | 2 |
| miR5269-3p | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| miR5271-5p | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 |
| miR5272-5p | 17 | 22 | 12 | 12 | 34 | 21 | 18 | 18 |
| miR5273-3p | 1 | 3 | 1 | 1 | 3 | 1 | 4 | 2 |
| miR5277-3p | 60 | 108 | 62 | 75 | 99 | 48 | 16 | 20 |
| miR5279-5p | 3 | 19 | 13 | 1 | 16 | 8 | 8 | 7 |
| miR5281-3p | 29 | 47 | 29 | 35 | 69 | 18 | 141 | 150 |
| miR5282-3p | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| miR5284-3p | 20 | 52 | 50 | 4 | 14 | 17 | 10 | 23 |
| miR5285-5p | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 3 |
| miR5286-3p | 2 | 0 | 2 | 1 | 3 | 2 | 3 | 2 |
| miR5287-3p | 6 | 10 | 14 | 8 | 9 | 4 | 17 | 19 |
| miR5289-3p | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| miR5290-3p | 0 | 5 | 1 | 1 | 2 | 1 | 2 | 6 |
| miR5291-3p | 0 | 1 | 1 | 0 | 3 | 1 | 0 | 1 |
| miR5292-3p | 16 | 35 | 21 | 6 | 34 | 21 | 53 | 82 |
| miR5294-3p | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| miR5295-3p | 9 | 29 | 13 | 3 | 15 | 9 | 7 | 6 |
| miR5296-3p | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| miR5297-3p | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 1 |
| miR5298-3p | 4 | 4 | 1 | 4 | 0 | 1 | 3 | 15 |
| miR5299-3p | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| miR5558-5p | 539 | 1938 | 1820 | 220 | 415 | 412 | 1103 | 1276 |
| miR5559-5p | 7 | 3 | 0 | 0 | 0 | 0 | 8 | 5 |
| miR5561-3p | 5 | 14 | 18 | 0 | 5 | 5 | 4 | 5 |
| miR5561-5p | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| miR5743-5p | 19 | 113 | 6 | 0 | 1 | 1 | 70 | 398 |
| miR5744-5p | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| miR5745-3p | 28 | 39 | 41 | 69 | 144 | 171 | 126 | 113 |
| miR5752-3p | 0 | 4 | 0 | 0 | 1 | 0 | 8 | 11 |
| miR5754-5p | 0 | 6 | 19 | 0 | 1 | 3 | 2 | 41 |
| miR7696-5p | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 |
| miR7696-3p | 174 | 95 | 253 | 40 | 138 | 184 | 1173 | 255 |
| miR7701-3p | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
MicroRNA profiles in alfalfa plants, roots and shoots
A total of 100 known miRNA families were identified from the small RNA libraries of the three alfalfa genotypes (Table 2). Of these, 21 families were represented by the highly conserved miRNAs, whereas the remaining 79 families could be considered as Medicago-specific miRNA families. The identification of these 79 miRNA families in alfalfa was based on their expression in M. truncatula (miRbase), therefore, these are conserved at least between M. truncatula and alfalfa.
Among the highly conserved miRNA families, miR166 was the most highly expressed family in seven of the eight samples that were surveyed in this study. The only exception to this trend was observed in the shoots of NECS-141 in which the miR2118 family was the most abundant followed by the miR166 family. The miRNA families, miR396 and miR2118 represents the second and third most abundantly expressed in the whole plants, while miR159 and miR396 were the second and third most highly expressed miRNAs in roots. Several additional miRNA families including miR398, miR160, miR168, miR319, miR408, miR1510 and miR2643 were also highly expressed but miR169, miR171, miR393, miR397 and miR395 were expressed at relatively very low levels (Table 2). On the other hand, miR159, miR156, miR319, miR398 miR1507 and miR1510 were highly expressed but miR164, miR169, miR172, miR393, miR397, miR399 and miR482 were expressed at very low levels in roots of these genotypes. Interestingly, miR160 was not sequenced from the roots of three alfalfa genotypes.
Overall, the conserved miRNA families such as the miR156, miR159, miR166, miR168, miR319, miR396, miR398 and miR408 were more highly expressed in the plants, roots and shoots of all three alfalfa genotypes. Among the legume-specific families, miR1507, miR1510, miR2118, miR2592, miR2643, miR5213, miR5232, miR5558 and miR7696 (Table 2) were also abundant in all tissues of alfalfa genotypes. Conversely, some conserved miRNA families represented by miR169 and miR393 recorded very low abundances in all samples. Other notable differences between roots and shoots include relatively low expression levels of miR160, miR167, and miR408 in roots compared to the shoots of alfalfa genotypes (Table 2).
Several miRNA families including miR482, miR1507, miR2118, miR4416 are conserved in M. truncatula, soybean, chickpea (miRBase). These miRNA families are known to regulate NBS-LRR genes that are involved in pathogen resistance. The miRNA-guided cleavage on the NBS-LRR genes initiates the generation of phasiRNAs [16, 18, 32]. In alfalfa, miR482, miR1507 and miR2118 were detected in all three tissues (Table 2), but not miR4416. Both miR2118 and miR1507 families were more abundantly expressed in all tissues and genotypes compared with miR482 family. Remarkably, miR2118 was the top most highly expressed miRNA family in shoots of NECS-141. By contrast, miR2118 levels were very low in roots of three alfalfa genotypes. On the other hand, miR1507 family displayed approximately similar levels in three tissues of alfalfa genotypes.
The miRNA-star sequences corresponding to the 12 of the 21 highly conserved miRNA families were also recovered from almost all libraries (Table 2). Additionally, miRNA-stars for the miR1510, miR4414, miR5208, and miR7696 were also detected. Furthermore, the miRNA-star expression levels for miR156, miR166 and miR168 were very high (Table 2). Intriguingly, like miR168, miR168 star levels differed greatly between different tissue. In shoots of NECS-141, miR168 star levels were slightly more than that of miR168, while both in whole plants and roots, the star levels were approximately half of the levels of miR168.
miRNA diversity in alfalfa compared with other legumes
Several miRNA families are specifically reported from the leguminous plants such as the M. truncatula, Glycine max, Lotus japonicus, Phaseolus vulgaris, Cicer arietinum, Vigna unguiculata and Acacia auriculiformis [14, 16, 18, 19, 32, 33]. These lineage-specific miRNAs include miR1507, miR1508, miR1509, miR1510, miR1512, miR1514, miR1520, miR1521, miR2118, miR2086, miR2109, miR2199, miR4414, miR5213, miR5232, and miR5234 among others (miRBase). The majority of these were reported from M. truncatula and soybean, since these legume species have been the subject of multiple studies exploring small RNAs. Most of these legume-specific miRNAs were also identified in alfalfa and a few of them including miR1507, miR1510, miR2118, miR2592, miR2643, miR5211, miR5213, miR5214, miR5232, miR5239, miR5277, miR5558, and miR7696 were specifically highly expressed in all three genotypes (Table 2).
Identification of novel miRNAs from alfalfa
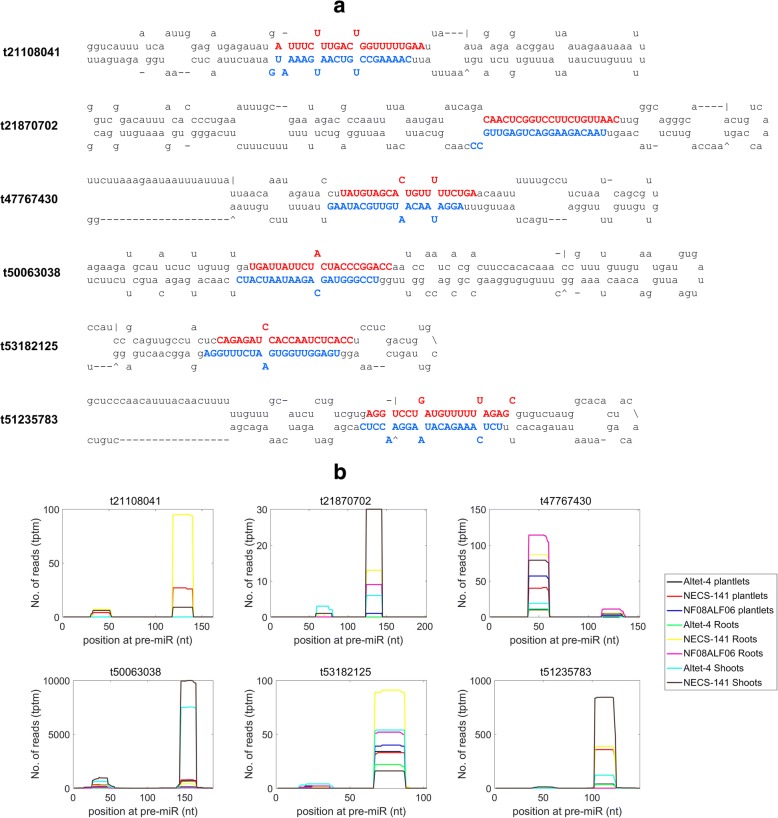
The sequencing of the small RNAs from multiple tissues of three different alfalfa genotypes would allow us to identify the novel miRNAs more confidently. Novel miRNA identification was dependent on sequencing of the miRNA complementary strand (miRNA-star) coupled with the predictable fold back structure for the primary miRNA transcript. Because a stable assembly of the tetraploid alfalfa genome sequence is not available, the small RNAs were mapped to the M. truncatula genome. Mapping of the small RNAs from the three alfalfa genotypes onto the M. truncatula genome enabled the identification of novel miRNAs more confidently because they have been sequenced from M. sativa and mapped on to the M. truncatula, suggesting their conservation between M. sativa and M. truncatula. Moreover, the novel miRNA identification in this study is more robust as it includes sequencing of these small RNAs from three different genotypes. We have identified a total of 17 novel miRNAs which have been sequenced from all of the three genotypes (Table 3 and Fig. 1). Among these, t50582913 was highly expressed followed by t50063038. In roots, t50582913 was highly expressed in NECS-141 and Altet-4 but not in NF08ALF06. In shoots, t50063038 was highly expressed followed by the t50582913 and t51235783.
Table 3.
Identified novel miRNAs based on sequencing both 5′ and 3′ reads and the most abundant ones that is marked in bold denotes potential novel miRNA based on their greater abundances
| miR-5p | miR-5p_seq | miR-3p | miR-3p_seq | Altet-4 Plants | NECS-141 Plants | NF08ALF06 Plants | Altet-4 Roots | NECS-141 Roots | NF08ALF06 Roots | Altet-4 Shoots | NECS-141 Shoots |
|---|---|---|---|---|---|---|---|---|---|---|---|
| t61680599 | UUUCUUUGACUGGUUUUUGAAU | t21108041 | CAAAAGCCUGUCAAUGAAAAUG | 0 | 31 | 0 | 0 | 312 | 0 | 0 | 32 |
| t46402976 | UAGCAUCAAGCGUCGCGUCGAU | t28372577 | CGACCCGAGGCUUAUGCGAUC | 115 | 97 | 145 | 81 | 479 | 229 | 335 | 315 |
| t59820880 | UUGGCAGAAUCACGGUGUGCC | t29809748 | CGGUGGCAUCGUGAUUUUGAC | 0 | 6 | 25 | 1 | 6 | 8 | 1 | 47 |
| t21870702 | CAACUCGGUCCUUCUGUUAAC | t44359413 | UAACAGAAGGACUGAGUUGCC | 0 | 11 | 3 | 1 | 41 | 12 | 24 | 103 |
| t62603216 | UUUUCAAGUUGGUCCCUUACG | t44814359 | UAAGGGACCAACUUGAAAACU | 77 | 178 | 196 | 7 | 240 | 107 | 529 | 899 |
| t8901469 | ACCUGGAGACAGAGAUGCAAU | t45832108 | UACGUCUCUGUCUUUCGGGUUG | 1 | 55 | 28 | 2 | 222 | 28 | 6 | 247 |
| t12927907 | AGGAUAACAAUGUUGCAUAAG | t47767430 | UAUGUAGCACUGUUUUUCUGA | 13 | 43 | 85 | 14 | 273 | 147 | 83 | 262 |
| t63076572 | UUUUUAGAUACAUUGAAUAAU | t47960370 | UAUUCAAUGUAUCUAAAAAG | 10 | 10 | 14 | 4 | 40 | 2 | 208 | 177 |
| t53501433 | UGAUUAUUCUACUACCCGGACC | t50063038 | UCCGGGUAGCAGAAUAAUCAUC | 350 | 371 | 78 | 45 | 371 | 18 | 17,057 | 20,494 |
| t12458129 | AGCGGUUGGUACAAUGCAAUAu | t50582913 | UCGCCUUGUACCAACCUACUGC | 544 | 915 | 0 | 123 | 1148 | 0 | 881 | 1453 |
| t40560414 | GGUCCUGAUGUUUUUUAGAGC | t51235783 | UCUCAAAGACAUAAGGAACCUC | 19 | 281 | 0 | 24 | 762 | 0 | 269 | 1655 |
| t55270980 | UGUCUUUAGCUUCCGAAACAa | t55621674 | UGUUCCGGUAGAUGAAGUCAC | 4 | 4 | 0 | 2 | 23 | 0 | 24 | 40 |
| t14211567 | AGUUAAUUGUGUUGCAUGAGUU | t57726911 | UUCAGCAACAUGAGUUAACUCA | 17 | 26 | 60 | 3 | 48 | 22 | 42 | 50 |
| t8194733 | ACAUUUUAGAUUGUUGAGGAA | t27568341 | CCUCAAUUAUCUAUUAUGUUU | 0 | 0 | 3 | 0 | 3 | 6 | 6 | 0 |
| t62313817 | UUUGUUAAACAUUUGUUUCC | t311560 | AAAACAAAUGUUUAGCUAAG | 0 | 6 | 0 | 0 | 15 | 1 | 0 | 12 |
| t55268921 | UGUCUUGGUUUCAAAAAGAAGu | t52170136 | UCUUUUUGCAAACCAACUCAAU | 4 | 19 | 13 | 1 | 29 | 4 | 9 | 56 |
| t51870988 | UCUUAUUUUCGACAUUGCAAAG | t59475847 | UUGCAGGUCGAGAAUAAAAUG | 19 | 99 | 71 | 1 | 9 | 1 | 353 | 1072 |
Fig. 1.
The predicted foldback structures using the novel miRNA precursor sequences. a The fold-back structures for six novel miRNAs. b The distribution of small RNA reads on the precursors of the novel miRNAs depicted in Fig. 1a
Identification of miRNA targets in alfalfa
Although the alfalfa is one of the important legumes agronomically, the genome sequencing and annotations are not available so far. Due to this, studies have utilized the well-studied and closely related M. truncatula genome annotations as a model for alfalfa studies. The nucleotide identity for some genes was greater than 97% between M. sativa and M. truncatula [34]). Thus, using M. truncatula transcript annotations can facilitate identification of miRNA targets in alfalfa. We used SeqTar algorithm (Zheng et al., 2012) to identify miRNA targets by allowing a maximum of 4 mismatches between miRNAs and their potential target transcripts.
Previous studies have revealed that conserved miRNAs are strongly associated with the regulation of genes that encode transcription factors [35]. These transcription factors in turn regulate key developmental processes and pathways in plants. Degradome sequencing has been very effective in identifying plant miRNA targets. Besides identifying the conserved targets, this approach can also identify non-conserved targets for the conserved miRNAs [28, 36, 37]. Degradome sequencing was used in this study to identify the cleaved mRNA fragments corresponding to the miRNA recognition sites in all three alfalfa genotypes. Approximately 30 million degradome reads were obtained from the transcripts of each of the alfalfa genotypes (Table 4) and these reads were analysed using SeqTar program. In total, we have identified 69 targets for 31 miRNA families that included 16 highly conserved families (Table 5). With respect to the conserved miRNAs, 33 targets for 16 conserved miRNA families were identified (Table 5). The known targets for miR162, miR165/166, miR398 and miR399 families were not identified in this study. Although miR165/166 family is the most abundantly expressed as scored from their read frequencies in almost all libraries but the cleaved fragments from the HD-Zip target transcripts were not recovered from degradome libraries of alfalfa genotypes.
Table 4.
Mapping of the reads obtained from the degradome libraries
| Database | Altet-4 | NECS-141 | NF08ALF06 | |||
|---|---|---|---|---|---|---|
| Total reads | Unique reads | Total reads | Unique reads | Total reads | Unique reads | |
| M. truncatula genome | 852,790 | 487,582 | 1,541,055 | 791,294 | 3,091,832 | 1,021230 |
| M. sativa genome | 1,488,681 | 957,866 | 2,691,763 | 1,435,659 | 4,591,130 | 1,877,041 |
| Cds | 770,970 | 426,278 | 1,436,059 | 727,330 | 2,928,098 | 933,425 |
| ncRNA | 231,076 | 22,907 | 186,813 | 18,014 | 1,305,681 | 36,585 |
| Repeats | 171,358 | 16,804 | 116,741 | 16,759 | 636,675 | 23,958 |
| Pre-miRBase | 34,631 | 837 | 35,979 | 1045 | 50,136 | 1192 |
| Total | 28,674,678 | 2,286,693 | 30,573,270 | 3,137,327 | 30,812,606 | 3,885,547 |
Table 5.
miRNA targets identified in the degradome libraries generated from three alfalfa genotypes. #Mis. is number of mismatches on the miRNA complementary site; Valid reads is Reads corresponding to the expected cleavage site; Total reads is Total reads mapped to the cDNA of the gene; Percent is Percent reads at the expected cleavage site
| genotypes | miRNA id# | Target gene | #Mis. | Valid reads | Total reads | Percent | Target gene annotation |
|---|---|---|---|---|---|---|---|
| Altet-4 | miR156e | Medtr7g028740.2 | 0 | 4 | 23 | 17.4 | squamosa promoter-binding-like protein |
| Altet-4 | miR156a | Medtr7g444860.1 | 0 | 2 | 28 | 7.1 | squamosa promoter-binding-like protein |
| Altet-4 | miR156a | Medtr3g099080.1 | 0 | 1 | 3 | 33.3 | squamosa promoter-binding 13A-like protein |
| Altet-4 | miR159b | Medtr8g042410.1 | 2.5 | 1 | 16 | 6.3 | MYB transcription factor |
| Altet-4 | miR160c | Medtr2g094570.3 | 1 | 4 | 21 | 19.1 | auxin response factor 1 |
| Altet-4 | miR164d | Medtr2g064470.1 | 1 | 2 | 34 | 5.9 | NAC transcription factor-like protein |
| Altet-4 | miR164d | Medtr8g058330.1 | 2 | 5 | 49 | 10.2 | protein transporter Sec61 subunit alpha-like protein |
| Altet-4 | miR167b-5p | Medtr8g079492.3 | 4 | 4 | 62 | 6.5 | auxin response factor 2 |
| Altet-4 | miR169e-5p | Medtr2g099490.2 | 2 | 1 | 20 | 5 | CCAAT-binding transcription factor |
| Altet-4 | miR171f | Medtr0092s0100.2 | 1.5 | 5 | 24 | 20.8 | GRAS family transcription regulator |
| Altet-4 | miR172a | Medtr4g094868.3 | 1 | 1 | 13 | 7.7 | AP2 domain transcription factor |
| Altet-4 | miR172a | Medtr5g016810.2 | 1 | 1 | 18 | 5.6 | AP2 domain transcription factor |
| Altet-4 | miR172a | Medtr2g093060.3 | 0 | 4 | 17 | 23.5 | AP2-like ethylene-responsive transcription factor |
| Altet-4 | miR319d-3p | Medtr2g078200.1 | 3 | 2 | 34 | 5.9 | TCP family transcription factor |
| Altet-4 | miR319d-3p | Medtr8g463380.1 | 3 | 2 | 7 | 28.6 | TCP family transcription factor |
| Altet-4 | miR393a | Medtr1g088950.1 | 1 | 11 | 83 | 13.3 | transport inhibitor response-like protein |
| Altet-4 | miR393a | Medtr7g083610.1 | 2 | 38 | 134 | 28.4 | transport inhibitor response 1 protein |
| Altet-4 | miR395j | Medtr1g102550.1 | 1 | 1 | 76 | 1.3 | ATP sulfurylase |
| Altet-4 | miR396b-5p | Medtr1g017490.2 | 3 | 47 | 100 | 47 | growth-regulating factor |
| Altet-4 | miR396b-5p | Medtr2g041430.3 | 3 | 5 | 12 | 41.7 | growth-regulating factor-like protein |
| Altet-4 | miR396b-5p | Medtr5g027030.1 | 3 | 5 | 15 | 33.3 | growth-regulating factor |
| Altet-4 | miR396a-5p | Medtr3g052060.1 | 2 | 1 | 1 | 100 | hypothetical protein |
| Altet-4 | miR398c | Medtr4g114870.1 | 3 | 8 | 23 | 34.8 | plastocyanin-like domain protein |
| Altet-4 | miR398a-3p | Medtr8g064810.1 | 3 | 5 | 36 | 13.9 | protein disulfide isomerase (PDI)-like protein |
| Altet-4 | miR408-3p | Medtr8g089110.1 | 3 | 3 | 9 | 33.3 | basic blue-like protein |
| Altet-4 | miR408-3p | Medtr8g007020.1 | 3.5 | 5 | 73 | 6.9 | plastocyanin-like domain protein |
| Altet-4 | miR408-3p | Medtr8g007035.1 | 3.5 | 5 | 123 | 4.1 | plastocyanin-like domain protein |
| Altet-4 | miR408-5p | Medtr3g074830.1 | 3.5 | 2 | 442 | 0.5 | phosphate-responsive 1 family protein |
| Altet-4 | miR1510a-5p | Medtr2g012770.1 | 1 | 1 | 5 | 20 | disease resistance protein (TIR-NBS-LRR class) |
| Altet-4 | miR2199 | Medtr7g080780.2 | 2 | 2 | 8 | 25 | helix loop helix DNA-binding domain protein |
| Altet-4 | miR2643a | Medtr3g010590.1 | 1 | 1 | 15 | 6.7 | F-box protein interaction domain protein |
| Altet-4 | miR2643a | Medtr6g053240.1 | 3 | 2 | 4 | 50 | F-box protein interaction domain protein |
| Altet-4 | miR4414a-5p | Medtr3g117120.1 | 4 | 3 | 84 | 3.6 | BZIP transcription factor bZIP124 |
| Altet-4 | miR5213-5p | Medtr6g084370.1 | 2 | 1 | 2 | 50 | disease resistance protein (TIR-NBS-LRR class) |
| Altet-4 | miR5213-5p | Medtr6g088245.1 | 3 | 1 | 5 | 20 | disease resistance protein (TIR-NBS-LRR class) |
| Altet-4 | miR5239 | Medtr3g018680.1 | 3 | 1 | 5 | 20 | F-box/RNI superfamily protein, putative |
| Altet-4 | miR5561-3p | Medtr2g045295.1 | 3 | 1 | 4 | 25 | hypothetical protein |
| Altet-4 | miR5752b | Medtr8g066820.1 | 4 | 9 | 423 | 2.1 | PLATZ transcription factor family protein | |
| Altet-4 | miR7696a-5p | Medtr1g072130.1 | 3 | 2 | 27 | 7.4 | PHD finger protein, putative |
| Altet-4 | miR7696c-3p | Medtr3g081480.1 | 3 | 2 | 21 | 9.5 | endoplasmic reticulum vesicle transporter |
| Altet-4 | miR7696d-5p | Medtr3g112250.1 | 3.5 | 8 | 36 | 22.2 | hypothetical protein |
| Altet-4 | miR7696c-3p | Medtr4g011600.2 | 3.5 | 1 | 26 | 3.9 | sulfate transporter-like protein |
| Altet-4 | miR7696c-3p | Medtr7g085650.4 | 3.5 | 1 | 6 | 16.7 | sulfate adenylyltransferase subunit 1/adenylylsulfate kinase |
| Altet-4 | miR7701-3p | Medtr6g011380.2 | 2 | 1 | 137 | 0.7 | SPFH/band 7/PHB domain membrane-associated family protein |
| NF08ALF06 | miR156e | Medtr7g028740.2 | 0 | 14 | 36 | 38.9 | squamosa promoter-binding-like protein |
| NF08ALF06 | miR156a | Medtr7g444860.1 | 0 | 1 | 144 | 0.7 | squamosa promoter-binding-like protein |
| NF08ALF06 | miR156h-3p | Medtr7g091370.1 | 3 | 1 | 11 | 9.1 | heat shock transcription factor |
| NF08ALF06 | miR159b | Medtr8g042410.1 | 2.5 | 4 | 30 | 13.3 | MYB transcription factor |
| NF08ALF06 | miR160c | Medtr2g094570.3 | 1 | 8 | 46 | 17.4 | auxin response factor 1 |
| NF08ALF06 | miR160d | Medtr1g064430.2 | 0.5 | 3 | 24 | 12.5 | auxin response factor 1 |
| NF08ALF06 | miR160d | Medtr3g073420.1 | 0.5 | 2 | 17 | 11.8 | auxin response factor, putative |
| NF08ALF06 | miR164d | Medtr2g064470.1 | 1 | 41 | 151 | 27.2 | NAC transcription factor-like protein |
| NF08ALF06 | miR164d | Medtr8g058330.1 | 2 | 5 | 115 | 4.4 | protein transporter Sec61 subunit alpha-like protein |
| NF08ALF06 | miR167b-5p | Medtr8g079492.3 | 4 | 9 | 133 | 6.8 | auxin response factor 2 |
| NF08ALF06 | miR167a | Medtr4g076020.1 | 3.5 | 5 | 77 | 6.5 | GRAS family transcription factor |
| NF08ALF06 | miR171f | Medtr0092s0100.2 | 1.5 | 60 | 115 | 52.2 | GRAS family transcription regulator |
| NF08ALF06 | miR172a | Medtr4g094868.3 | 1 | 1 | 45 | 2.2 | AP2 domain transcription factor |
| NF08ALF06 | miR172a | Medtr5g016810.2 | 1 | 1 | 84 | 1.2 | AP2 domain transcription factor |
| NF08ALF06 | miR172a | Medtr2g093060.3 | 0 | 4 | 35 | 11.4 | AP2-like ethylene-responsive transcription factor |
| NF08ALF06 | miR172a | Medtr4g061200.4 | 1 | 1 | 28 | 3.6 | AP2-like ethylene-responsive transcription factor |
| NF08ALF06 | miR172a | Medtr7g100590.1 | 1 | 2 | 17 | 11.8 | AP2 domain transcription factor |
| NF08ALF06 | miR319d-3p | Medtr2g078200.1 | 3 | 2 | 126 | 1.6 | TCP family transcription factor |
| NF08ALF06 | miR319d-3p | Medtr8g463380.1 | 3 | 2 | 48 | 4.2 | TCP family transcription factor |
| NF08ALF06 | miR393a | Medtr1g088950.1 | 1 | 54 | 268 | 20.2 | transport inhibitor response-like protein |
| NF08ALF06 | miR393a | Medtr7g083610.1 | 2 | 472 | 771 | 61.2 | transport inhibitor response 1 protein |
| NF08ALF06 | miR393a | Medtr8g098695.2 | 4 | 1 | 46 | 2.2 | transport inhibitor response 1 protein |
| NF08ALF06 | miR396b-5p | Medtr1g017490.2 | 3 | 423 | 742 | 57 | growth-regulating factor |
| NF08ALF06 | miR396b-5p | Medtr2g041430.3 | 3 | 30 | 75 | 40 | growth-regulating factor-like protein |
| NF08ALF06 | miR396b-5p | Medtr5g027030.1 | 3 | 10 | 42 | 23.8 | growth-regulating factor |
| NF08ALF06 | miR396a-5p | Medtr3g011560.1 | 3 | 1 | 3 | 33.3 | TNP1 |
| NF08ALF06 | miR396a-5p | Medtr3g052060.1 | 2 | 3 | 11 | 27.3 | hypothetical protein |
| NF08ALF06 | miR396a-5p | Medtr8g017000.1 | 3 | 1 | 2 | 50 | Ulp1 protease family, carboxy-terminal domain protein |
| NF08ALF06 | miR398c | Medtr4g114870.1 | 3 | 14 | 49 | 28.6 | plastocyanin-like domain protein |
| NF08ALF06 | miR398a-3p | Medtr8g064810.1 | 3 | 8 | 44 | 18.2 | protein disulfide isomerase (PDI)-like protein |
| NF08ALF06 | miR408-3p | Medtr8g089110.1 | 3 | 8 | 34 | 23.5 | basic blue-like protein |
| NF08ALF06 | miR408-3p | Medtr8g007020.1 | 3.5 | 10 | 375 | 2.7 | plastocyanin-like domain protein |
| NF08ALF06 | miR408-3p | Medtr8g007035.1 | 3.5 | 10 | 675 | 1.5 | plastocyanin-like domain protein |
| NF08ALF06 | miR408-5p | Medtr3g074830.1 | 3.5 | 27 | 948 | 2.9 | phosphate-responsive 1 family protein |
| NF08ALF06 | miR482-5p | Medtr1g064430.2 | 3.5 | 1 | 24 | 4.2 | auxin response factor 1 |
| NF08ALF06 | miR530 | Medtr3g072110.1 | 2.5 | 3 | 102 | 2.9 | transmembrane amino acid transporter family protein |
| NF08ALF06 | miR1507–3p | Medtr8g036195.1 | 2 | 4 | 9 | 44.4 | NBS-LRR type disease resistance protein |
| NF08ALF06 | miR1510a-5p | Medtr7g108860.4 | 3.5 | 21 | 1061 | 2 | CS domain protein |
| NF08ALF06 | miR2199 | Medtr7g080780.2 | 2 | 1 | 26 | 3.9 | helix loop helix DNA-binding domain protein |
| NF08ALF06 | miR2643a | Medtr6g053240.1 | 3 | 25 | 33 | 75.8 | F-box protein interaction domain protein |
| NF08ALF06 | miR4414a-5p | Medtr3g117120.1 | 4 | 8 | 260 | 3.1 | BZIP transcription factor bZIP124 |
| NF08ALF06 | miR5037c | Medtr4g070550.1 | 3 | 2 | 44 | 4.6 | F-box protein interaction domain protein |
| NF08ALF06 | miR5213-5p | Medtr4g014580.1 | 1.5 | 3 | 31 | 9.7 | TIR-NBS-LRR class disease resistance protein |
| NF08ALF06 | miR5238 | Medtr3g077740.2 | 2.5 | 1 | 259 | 0.4 | pantothenate kinase |
| NF08ALF06 | miR5239 | Medtr3g018680.1 | 3 | 4 | 43 | 9.3 | F-box/RNI superfamily protein, putative |
| NF08ALF06 | miR5561-3p | Medtr2g045295.1 | 3 | 1 | 12 | 8.3 | hypothetical protein |
| NF08ALF06 | miR5752a | Medtr8g066820.1 | 4 | 13 | 936 | 1.4 | PLATZ transcription factor family protein |
| NF08ALF06 | miR7696a-5p | Medtr1g072130.1 | 3 | 4 | 259 | 1.5 | PHD finger protein, putative |
| NF08ALF06 | miR7696c-3p | Medtr3g081480.1 | 3 | 2 | 46 | 4.4 | endoplasmic reticulum vesicle transporter |
| NF08ALF06 | miR7696c-5p | Medtr7g076830.1 | 3 | 3 | 103 | 2.9 | DEAD-box ATP-dependent RNA helicase-like protein |
| NF08ALF06 | miR7696d-5p | Medtr3g112250.1 | 3.5 | 5 | 30 | 16.7 | hypothetical protein |
| NF08ALF06 | miR7696c-3p | Medtr4g011600.2 | 3.5 | 1 | 103 | 1 | sulfate transporter-like protein |
| NF08ALF06 | miR7696c-3p | Medtr7g085650.4 | 3.5 | 2 | 10 | 20 | sulfate adenylyltransferase subunit 1/adenylylsulfate kinase |
| NF08ALF06 | miR7701-3p | Medtr3g108910.1 | 2.5 | 2 | 375 | 0.5 | hypothetical protein |
| NF08ALF06 | miR7701-3p | Medtr6g011380.2 | 2 | 2 | 86 | 2.3 | SPFH/band 7/PHB domain membrane-associated family protein |
| NCES-141 | miR156e | Medtr7g028740.2 | 0 | 18 | 46 | 39.1 | squamosa promoter-binding-like protein |
| NCES-141 | miR156a | Medtr7g444860.1 | 0 | 4 | 101 | 4 | squamosa promoter-binding-like protein |
| NCES-141 | miR156a | Medtr8g096780.1 | 0 | 1 | 11 | 9.1 | squamosa promoter-binding 13A-like protein |
| NCES-141 | miR156a | Medtr3g085180.1 | 1 | 1 | 2 | 50 | squamosa promoter-binding-like protein |
| NCES-141 | miR156h-3p | Medtr7g091370.1 | 3 | 2 | 5 | 40 | heat shock transcription factor |
| NCES-141 | miR159b | Medtr8g042410.1 | 2.5 | 3 | 36 | 8.3 | MYB transcription factor |
| NCES-141 | miR160c | Medtr2g094570.3 | 1 | 12 | 37 | 32.4 | auxin response factor 1 |
| NCES-141 | miR164d | Medtr2g064470.1 | 1 | 33 | 100 | 33 | NAC transcription factor-like protein |
| NCES-141 | miR164d | Medtr8g058330.1 | 2 | 14 | 119 | 11.8 | protein transporter Sec61 subunit alpha-like protein |
| NCES-141 | miR167b-5p | Medtr8g079492.3 | 4 | 10 | 101 | 9.9 | auxin response factor 2 |
| NCES-141 | miR167a | Medtr4g076020.1 | 3.5 | 4 | 45 | 8.9 | GRAS family transcription factor |
| NCES-141 | miR167b-3p | Medtr4g124900.2 | 3.5 | 1 | 154 | 0.7 | auxin response factor 2 |
| NCES-141 | miR168a | Medtr6g477980.2 | 4 | 2 | 245 | 0.8 | argonaute protein 1A |
| NCES-141 | miR171f | Medtr0092s0100.2 | 1.5 | 36 | 70 | 51.4 | GRAS family transcription regulator |
| NCES-141 | miR172a | Medtr4g094868.3 | 1 | 2 | 50 | 4 | AP2 domain transcription factor |
| NCES-141 | miR172a | Medtr5g016810.2 | 1 | 2 | 56 | 3.6 | AP2 domain transcription factor |
| NCES-141 | miR172a | Medtr2g093060.3 | 0 | 1 | 19 | 5.3 | AP2-like ethylene-responsive transcription factor |
| NCES-141 | miR172a | Medtr4g061200.4 | 1 | 3 | 32 | 9.4 | AP2-like ethylene-responsive transcription factor |
| NCES-141 | miR319d-3p | Medtr2g078200.1 | 3 | 1 | 55 | 1.8 | TCP family transcription factor |
| NCES-141 | miR319d-3p | Medtr8g463380.1 | 3 | 1 | 26 | 3.9 | TCP family transcription factor |
| NCES-141 | miR393a | Medtr1g088950.1 | 1 | 38 | 222 | 17.1 | transport inhibitor response-like protein |
| NCES-141 | miR393a | Medtr7g083610.1 | 2 | 337 | 539 | 62.5 | transport inhibitor response 1 protein |
| NCES-141 | miR395j | Medtr1g102550.1 | 1 | 1 | 163 | 0.6 | ATP sulfurylase |
| NCES-141 | miR396b-5p | Medtr1g017490.2 | 3 | 201 | 352 | 57.1 | growth-regulating factor |
| NCES-141 | miR396b-5p | Medtr5g027030.1 | 3 | 6 | 16 | 37.5 | growth-regulating factor |
| NCES-141 | miR396b-5p | Medtr8g020560.1 | 3 | 1 | 7 | 14.3 | growth-regulating factor-like protein |
| NCES-141 | miR396a-5p | Medtr3g011560.1 | 3 | 1 | 1 | 100 | TNP1 |
| NCES-141 | miR396a-5p | Medtr8g017000.1 | 3 | 1 | 1 | 100 | Ulp1 protease family, carboxy-terminal domain protein |
| NCES-141 | miR397-5p | Medtr7g062310.1 | 1.5 | 2 | 4 | 50 | laccase/diphenol oxidase family protein |
| NCES-141 | miR398c | Medtr4g114870.1 | 3 | 8 | 21 | 38.1 | plastocyanin-like domain protein |
| NCES-141 | miR398a-3p | Medtr8g064810.1 | 3 | 47 | 89 | 52.8 | protein disulfide isomerase (PDI)-like protein |
| NCES-141 | miR398c | Medtr5g089180.1 | 3 | 4 | 19 | 21.1 | hypothetical protein |
| NCES-141 | miR408-3p | Medtr8g089110.1 | 3 | 9 | 18 | 50 | basic blue-like protein |
| NCES-141 | miR408-3p | Medtr8g007020.1 | 3.5 | 7 | 209 | 3.4 | plastocyanin-like domain protein |
| NCES-141 | miR408-3p | Medtr8g007035.1 | 3.5 | 8 | 381 | 2.1 | plastocyanin-like domain protein |
| NCES-141 | miR408-5p | Medtr3g074830.1 | 3.5 | 14 | 703 | 2 | phosphate-responsive 1 family protein |
| NCES-141 | miR482-3p | Medtr5g027900.1 | 2.5 | 1 | 19 | 5.3 | disease resistance protein (CC-NBS-LRR class) family protein |
| NCES-141 | miR530 | Medtr3g072110.1 | 2.5 | 1 | 119 | 0.8 | transmembrane amino acid transporter family protein |
| NCES-141 | miR1510a-5p | Medtr7g108860.4 | 3.5 | 17 | 746 | 2.3 | CS domain protein |
| NCES-141 | miR2643a | Medtr3g010620.1 | 1 | 2 | 72 | 2.8 | F-box protein interaction domain protein |
| NCES-141 | miR4414a-5p | Medtr3g117120.1 | 4 | 2 | 134 | 1.5 | BZIP transcription factor bZIP124 |
| NCES-141 | miR5037c | Medtr4g070550.1 | 3 | 1 | 36 | 2.8 | F-box protein interaction domain protein |
| NCES-141 | miR5213-5p | Medtr6g084370.1 | 2 | 1 | 5 | 20 | disease resistance protein (TIR-NBS-LRR class) |
| NCES-141 | miR5213-5p | Medtr4g014580.1 | 1.5 | 1 | 18 | 5.6 | TIR-NBS-LRR class disease resistance protein |
| NCES-141 | miR5213-5p | Medtr6g088245.1 | 3 | 1 | 7 | 14.3 | disease resistance protein (TIR-NBS-LRR class) |
| NCES-141 | miR5238 | Medtr3g077740.2 | 2.5 | 1 | 151 | 0.7 | pantothenate kinase |
| NCES-141 | miR5561-3p | Medtr2g045295.1 | 3 | 1 | 9 | 11.1 | hypothetical protein |
| NCES-141 | miR5752b | Medtr8g066820.1 | 4 | 8 | 765 | 1.1 | PLATZ transcription factor family protein |
| NCES-141 | miR7696a-5p | Medtr1g072130.1 | 3 | 2 | 135 | 1.5 | PHD finger protein, putative |
| NCES-141 | miR7696c-5p | Medtr7g076830.1 | 3 | 5 | 78 | 6.4 | DEAD-box ATP-dependent RNA helicase-like protein |
| NCES-141 | miR7696d-5p | Medtr3g112250.1 | 3.5 | 9 | 44 | 20.5 | hypothetical protein |
| NCES-141 | miR7696c-3p | Medtr4g011600.2 | 3.5 | 1 | 124 | 0.8 | sulfate transporter-like protein |
| NCES-141 | miR7696c-3p | Medtr7g085650.4 | 3.5 | 1 | 10 | 10 | sulfate adenylyltransferase subunit 1/adenylylsulfate kinase |
| NCES-141 | miR7701-3p | Medtr3g108910.1 | 2.5 | 2 | 444 | 0.5 | hypothetical protein |
The identified miRNA targets in all three genotypes include mainly transcription factors. Specifically, five members of the squamosa promoter-binding-like protein (SPL) targeted by the miR156 family, five members of the auxin response factors targeted by both miR160 and miR167 families, five members of the apetala2 (AP2)-domain containing transcription factors, four members of the growth-regulating factor (GRFs) family targeted by miR396, two members of the TCP family transcription factors targeted by miR319, and, a NAC domain-containing transcription factor-like protein (NAC) targeted by miR164 [35]. Additionally, transcripts encoding Argonaute targeted by miR168, laccase targeted by miR397, and three plantacyanin containing proteins targeted by miR408 were also identified. Although evidence indicates that that miR398 targets Cu/Zn superoxide dismutases and a copper chaperone for the superoxide dismutases (CCS) in plants [28, 38] these relationships were not apparent in the data from this study. On the other hand, we have identified three potentially non-conserved targets (plastocyanin, protein disulphide isomerase and a hypothetical protein) for miR398 in three alfalfa genotypes. In addition to the GRFs, our analyses revealed potential non-conserved targets for miR396 including TNP1, Ulp1 protease and hypothetical proteins (Table 5).
The analyses of legume-specific miRNAs and their targets have revealed an interesting miRNA: target networks between the miRNAs and the NBS-LRR genes [14, 16, 18, 32]. In this study, we identified NBS-LRR disease resistance genes as targets for four different miRNA families including miR482, miR1507, miR1510 and miR5213 in alfalfa (Table 5).
Degradome analyses has also been utilized to identify potential targets for several non-conserved miRNAs or miRNAs that are present only in closely related species such as the M. truncatula. To increase the confidence in identification of targets for the non-conserved miRNAs that are usually expressed at low abundances and the cleavage frequencies on those targets are relatively low, we considered as ‘targets’ only those for which the cleavages were detected at least in two of the three alfalfa genotypes. The transcripts for Medtr6g053240.1 (F-box protein interaction domain protein) had a cleavage frequency of approximately 75% and were targeted by the miR2643 in NF08ALF06 genotype. Additionally, two other F-box protein interaction domain protein genes were also identified as targets for miR2643 in alfalfa genotypes (Table 5). These results suggest that the F-box protein interaction domain protein family are regulated by this potential legume-specific miRNA. Another notable observation is that 6 different genes identified as potential targets for miR7696, and the cleavage frequency of a particular target gene (hypothetical protein, Medtr3g112250.1) was more abundant in all three alfalfa genotypes (Table 5).
Because some of the miRNA-stars are also highly expressed, we scrutinized the degradome reads for potential cleavages on the transcripts that are complementary to the miRNA-stars. This analysis has identified potential targets for at least four conserved miRNAs. Specifically, miR156-star targets a heat shock transcription factor, miR164-star targets a protein transporter Sec61 subunit alpha-like protein, miR167-star targets a GRAS family transcription factor, and, miR482-star targets an auxin response factor 1 in alfalfa (Table 5).
The confirmed targets of conserved miRNAs are known to regulate diverse developmental processes in the lifecycle of plants. For example, the SPL transcription factors (target of miR156) which regulate the transition from juvenile to adult phase of the life cycle in land plants [39]. Auxin receptors (TIR1 proteins) and ARFs targeted by miR393 and miR160, miR167, are components of the auxin signalling pathway that regulates several aspects of plant growth and development. The roles of NAC factors (targeted by miR164) include shoot meristem initiation and later root formation in Arabidopsis [40, 41]. Similarly, TCP family transcription factors have several different roles including regulating leaf morphogenesis [42, 43]. In Arabidopsis, seven out of nine GRFs are known targets for miR396 [44], and we have identified four GRFs as targets for miR396 in alfalfa (Table 5). By interacting with its coactivators called GRF-interacting factors (GIFs), this regulatory network (miR396-GRFs-GIFs) regulate leaf size, leaf growth and senescence in Arabidopsis [44]. The known targets for miR397 include laccase, which is involved in oxidative polymerization of lignin in plants [45]. Similarly, miR408 is targeting a family of plantacyanins, which could function in shuttling electron-transfer between proteins [46, 47].
The miR398 family is known to target CSDs and a copper chaperone for superoxide dismutase (CCS) genes in plants [28, 38]. In this study, we have identified plastocyanin-domain like proteins (plastocyanin is an essential electron carrier which shuttles the electrons between cytochrome b6f and PS I) represents a novel target for miR398. Protein disulphide isomerase (PDI) is a member of a family of dithiol/disulfide oxidoreductases, the thioredoxin superfamily, which functions in the formation of disulphide linkage between the cysteine residues for proper protein folding [48]. Our degradome analyses confirms that PDI represents a novel target for miR398 in alfalfa (Table 5). The other confirmed miRNA target transcripts include Leucine rich repeat resistance (LRR) proteins (TIR-NBS-LRR and CC-NBS-LRR) that play important roles in plant pathogen recognition and activation of plant innate immune responses [14, 16, 18, 32]. Yet another interesting target include the F-box protein interaction domain proteins that are regulated by miR2643, one of the very abundantly expressed miRNA in alfalfa.
Conclusions
The analyses of small RNA libraries from the whole plants, shoots and roots resulted in the identification of 100 miRNA families that included highly conserved miRNAs as well as miRNAs that are at least conserved between M. truncatula and alfalfa. The conserved miRNA profiles share some similarities and a few differences between genotypes and types of tissues (roots and shoots). The tissue-specific profiles were used to identify miRNAs that are highly abundant as well as those miRNAs that are expressed at low levels. Additionally, 17 novel miRNAs with varying levels of expression were also identified in alfalfa. The present study also reports identification of 69 targets for 31 miRNA families. In addition to the conserved targets for conserved miRNAs, a few non-conserved targets such as the PDI for miR398 were confirmed. Similarly, miR2643 is targeting three transcripts encoding F-box protein interaction domain containing proteins in alfalfa. In summary, the results from this study have increased our understanding of miRNAs and miRNA-mediated gene regulation in alfalfa that could result in potential tangible targets for practical applications in alfalfa and related legume species to increase biomass yield and address abiotic and biotic limitations to agricultural productivity.
Acknowledgements
Not applicable.
Funding
This research was funded by the Noble Research Institute and Forage Genetics International, a hatch grant from NIFA-0229360 (OKL02844) to RS and MM, and the National Natural Science Foundation of China (numbers 31460295 and 31760314) to YZ. This work was also partially supported by the Neustadt-Sarkeys Distinguished Professorship to RS. Publication costs are funded by the Oklahoma Agricultural Experiment Station, Oklahoma State University, Stillwater.
Availability of data and materials
The small RNA and degradome datasets generated and analyzed in the present study are available in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession number GSE119460 available at: https://www.ncbi.nlm.nih.gov/geo/query/511acc.cgi?acc=GSE119460.
About this supplement
This article has been published as part of BMC Genomics Volume 19 Supplement 10, 2018: Proceedings of the 29th International Conference on Genome Informatics (GIW 2018): genomics. The full contents of the supplement are available online at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-19-supplement-10.
Authors’ contributions
RS and MM conceived the idea and designed the study. CM and TH cultured the plants used in this study; RP isolated the RNA from samples and generated the small RNA libraries; YZ, SR, QW, SA and RS analyzed the small RNA and degradome libraries; RP, SA and RS wrote the manuscript; MM edited the manuscript. All authors reviewed and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Robert Pokoo, Email: robertpokoo@gmail.com.
Shuchao Ren, Email: 18469100972@163.com.
Qingyi Wang, Email: wqy19941211@163.com.
Christy M. Motes, Email: cmmotes@noble.org
Timothy D. Hernandez, Email: tdhernandez@noble.org
Sayvan Ahmadi, Email: sayvan@okstate.edu.
Maria J. Monteros, Email: mjmonteros@noble.org
Yun Zheng, Email: zhengyun5488@gmail.com.
Ramanjulu Sunkar, Phone: 405-744-8496, Email: ramanjulu.sunkar@okstate.edu.
References
- 1.Sheaffer CC, Martin NP, Lamb JFS, Cuomo GR, Grimsbo Jewett J, Quering SR. Leaf and stem properties of alfalfa entries. Agron J. 2000;92(4):733–739. doi: 10.2134/agronj2000.924733x. [DOI] [Google Scholar]
- 2.Doole GJ, Pannell DJ. Role and value of including lucerne (Medicago sativa L.) phases in crop rotations for the management of herbicide-resistant Lolium rigidum in Western Australia. Crop Prot. 2008;27:497–504. doi: 10.1016/j.cropro.2007.07.018. [DOI] [Google Scholar]
- 3.Latta RA, Blacklow LJ, Cocks PS. Comparative soil water, pasture production, and crop yields in phase farming systems with lucerne and annual pasture in Western Australia. Aust J Agric Res. 2001;52:295–303. doi: 10.1071/AR99168. [DOI] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Sunkar R, Zhu JK. MicroRNAs and short-interfering RNAs in plants. J Integr Plant Biol. 2007;49:817–826. doi: 10.1111/j.1744-7909.2007.00499.x. [DOI] [Google Scholar]
- 6.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Ann Rev Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- 8.Chen X. Small RNAs and their roles in plant development. Ann Rev Cell and Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunkar R. MicroRNAs with macro effects on plant stress responses. Semin Cell Dev Biol. 2010;21(8):805–811. doi: 10.1016/j.semcdb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Sunkar R, Jagadeeswaran G. In silico identification of conserved miRNAs in large number of diverse plant species. BMC Plant Biol. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axtell MJ, Snyder JP, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics. 2008;9(1):593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagadeeswaran G, Zheng Y, Li YF, Shukla LI, Matts J, Hoyt P, Sunkar R. Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. 2009;184(1):85–98. doi: 10.1111/j.1469-8137.2009.02915.x. [DOI] [PubMed] [Google Scholar]
- 15.Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, et al. Genome-wide Medicagotruncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009;21(9):2780–2796. doi: 10.1105/tpc.109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai J, Jeong D, DePaoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, Stacey G, Cook DR, Green PJ, Sherrier DJ, Meyers BC. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans- acting siRNAs. Genes Dev. 2001;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi X, Yang Q, Chen X, Wang J, Pan L, Chen M, He Y, Laiang X, Yu S. Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS One. 2011;6(11):e27530. doi: 10.1371/journal.pone.0027530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava S, Zheng Y, Kudapa H, Jagadeeswaran G, Hivrale V, Varshney RK, Sunkar R. High throughput sequencing of small RNA component of leaves and inflorescence revealed conserved and novel miRNAs as well as phasiRNA loci in chickpea. Plant Sci. 2015;235:46–57. doi: 10.1016/j.plantsci.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y, Hivrale V, Valliyodan B, Lelandais-Brière C, Farmer AD, May GD, Crespi M, Nguyen HT, Sunkar R. Small RNA profiles in soybean primary root tips under water deficit. BMC Syst Biol. 2016;10(5):126. doi: 10.1186/s12918-016-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan W, Zhang S, Du H, Sun X, Shi Y, Wang C. Genome-wide identification of different dormant Medicago sativa L. MicroRNAs in response to fall dormancy. PLoS One. 2014;9(12):e114612. doi: 10.1371/journal.pone.0114612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Wan L, Bi W, Wan X, Li Z, Cao J, Tong Z, Xu H, He F, Lo X. Identification of drought-responsive microRNAs from roots and leaves of alfalfa by high-throughput sequencing. Genes. 2017;8:119. doi: 10.3390/genes8040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Xu H, Li Y, Wan X, Ma Z, Cao J, Li Z, He F, Wang Y, Wan L, Tong Z, Li X. Analysis of physiological and miRNA responses to pi deficiency in alfalfa (Medicago sativa L.) Plant Mol Biol. 2018;96:473–492. doi: 10.1007/s11103-018-0711-3. [DOI] [PubMed] [Google Scholar]
- 23.Monteros MJ, Tang H, Ramaraj T, Devitt NP, Cameron CT, Bharti AK, Mudge J, Farmer AD, Motes CM, Town CD, Brummer EC, Udvardi M. Progress in sequencing the genome of tetraploid alfalfa (Medicago sativa L) San Diego: Plant and Animal Genome Conference. 2015. Jan 10–14; 2015. [Google Scholar]
- 24.Khu DM, Reyno R, Brummer EC, Bouton JH, Monteros MJ. Identification of aluminum tolerance QTLs in tetraploid alfalfa (Medicago sativa L.) Crop Sci. 2013;53:148–163. doi: 10.2135/cropsci2012.03.0181. [DOI] [Google Scholar]
- 25.Khu DM, Reyno R, Brummer EC, Monteros MJ. Screening methods for aluminum tolerance in alfalfa (Medicago sativa L.) Crop Sci. 2012;52:161–167. doi: 10.2135/cropsci2011.05.0256. [DOI] [Google Scholar]
- 26.Jagadeeswaran J, Nimmakayala P, Zheng Y, Gowdu K, Reddy U, Sunkar R. Characterization of the small RNA component of leaves and fruits from four different cucurbit species. BMC Genomics. 2012;13:329. doi: 10.1186/1471-2164-13-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Sunkar R. Transcriptome-wide identification of microRNA targets in rice. The Plant J. 2010;62(5):742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Y, Li Y-F, Sunkar R, Zhang W. SeqTar: an effective method for identifying MicroRNA guided cleavage sites from Degradome of Polyadenylated transcripts in plants. Nucleic Acids Res. 2012;40:e28. doi: 10.1093/nar/gkr1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8(1):25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Zheng Y, Jagadeeswaran G, Sunkar R. Characterization of small RNAs and their target genes in wheat seedlings using sequencing-based approaches. Plant Sci. 2013;203:17–24. doi: 10.1016/j.plantsci.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci U S A. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohli D, Joshi G, Deokar AA, Bhardwaj AR, Agarwal M, Katiyar-Agarwal S, Srinivasan R, Jain PK. Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLoS One. 2014;9(10):e108851. doi: 10.1371/journal.pone.0108851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesfaye M, Silverstein K, Bucciarelli B, Samac CD, Vance CP. The Affymetrix Medicago GeneChip® array is applicable for transcript analysis of alfalfa (Medicago sativa). Funct Plant Biol. 2006;33:783–8. [DOI] [PubMed]
- 35.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 36.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18(10):758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.German MA, Pillay M, Jeong D-HH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 38.Sunkar R, Kapoor A, Zhu JK. Post transcriptional induction of two cu/Zn superoxide dismutase genes in Arabidopsis is mediated by down-regulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poethig RS. Small RNAs and developmental timing in plants. Curr Opin Genet Dev. 2009;19:374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131:4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- 42.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 43.Koyama T, Sato F, Ohme-Takagi M. Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 2017;175(2):874–885. doi: 10.1104/pp.17.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inze D, Rodriguez RE, Palatnik JF. Post-transcriptional control of GRF transcription factors by microRNA396 and GIF co-activators affects leaf size and longevity. Plant J. 2014;79:413–426. doi: 10.1111/tpj.12567. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Q, Nakashima J, Chen F, Yin YB, Fu CX, Yun JF, Shao H, Wang XQ, Wang ZY, Dixon RA. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25:3976–3987. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nersissian AM, Immoos C, Hill MG, Hart PJ, Williams G, Herrmann RG, Valentine JS. Uclacyanins, stellacyanins, and plantacyanins ape distinct subfamilies of phytocyanins: plant-specific mononuclear blue copper proteins. Protein Sci. 1998;7:1915–1929. doi: 10.1002/pro.5560070907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma C, Burd S, Lers A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015;84:169–187. doi: 10.1111/tpj.12999. [DOI] [PubMed] [Google Scholar]
- 48.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Boiochem Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The small RNA and degradome datasets generated and analyzed in the present study are available in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession number GSE119460 available at: https://www.ncbi.nlm.nih.gov/geo/query/511acc.cgi?acc=GSE119460.