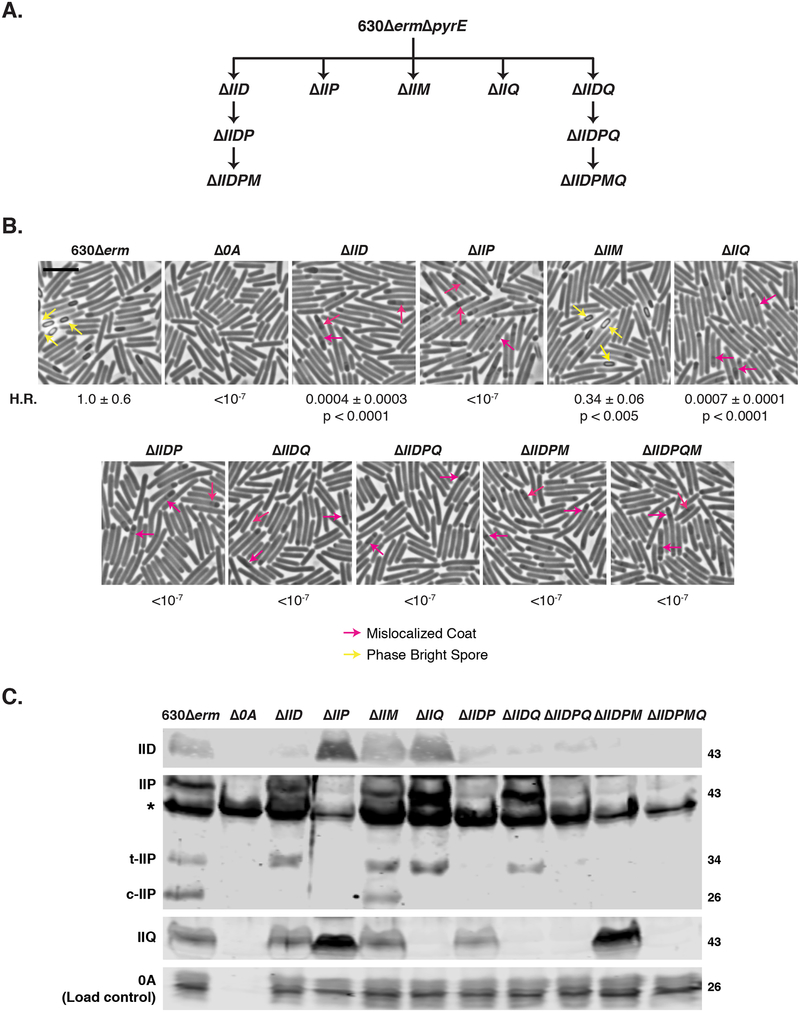
Fig. 2. Differential requirements for engulfment-related proteins during C. difficile spore formation.
(A) Family tree of putative engulfment mutants. 630ΔermΔpyrE was the parental strain for making first-generation single gene deletions as well as the spoIID-spoIIQ double gene deletion mutant. Additional multiple gene deletions were constructed using either ΔIID and ΔIIDQ as the parental strains. (B) Phase-contrast microscopy analyses of wild type 630Δerm, Δspo0A (Δ0A), and mutants lacking putative engulfment regulators after 17 hrs of sporulation. Yellow arrows mark example phase-bright forespores, while pink arrows demarcate regions suspected to be mislocalized coat based on previous studies (Fimlaid et al., 2015b, Ribis et al., 2017). Heat resistance (H.R.) efficiencies were determined from 20–24 sporulating cultures and represent the mean and standard deviation for a given strain relative to wild type based on a minimum of three independent biological replicates. Statistical significance relative to wild type was determined using a one-way ANOVA and Tukey’s test. Scale bars represent 5 μm. The limit of detection of the assay is 10−6. (C) Western blot analyses of IID, IIP, and IIQ in mutants lacking putative engulfment regulators. Three separate isoforms were detected for IIP in wild type: full-length (IIP), truncated IIP (t-IIP), and cleaved IIP (c-IIP). The predicted MW of IIP is 38 kDa, and the MW of IIP lacking its signal peptide is 35 kDa. The asterisk denotes a non-specific protein recognized by the polyclonal anti-IIP antibodies. A faint cross-reacting band is also visible with the IID antibody. Spo0A was used as a proxy for measuring sporulation induction (Dembek et al., 2017, Putnam et al., 2013). The western blots shown are representative of the results of three independent biological replicates.