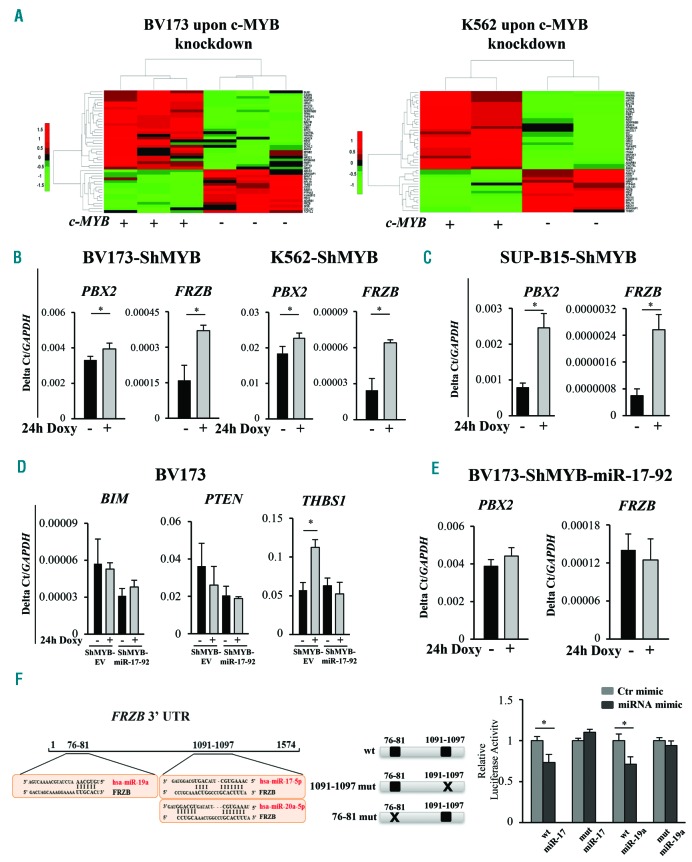
Figure 4.
Transcriptional analysis and evaluation of mRNA expression levels of miR-17-92 cluster target genes. (A) Unsupervised hierarchical clustering of common deregulated genes from gene expression analysis of parental and MYB-silenced BV173 and K562 cells. (B and C) qRT-PCR of PBX2 and FRZB expression levels upon MYB knockdown [24 hours (h)] of the indicated Philadelphia-positive (Ph+) ShMYB cell lines. Results are mean of three experiments. Error bars indicate Standard Error of Mean (SEM). (D) Analysis of mRNA expression levels, using SYBR Green-based qRT-PCR, of BIM, PTEN and THBS1 in untreated and Doxy-treated BV173-ShMYB-Empty Vector (EV) and ShMYB-miR-17-92 cells. Results are mean of three experiments. Error bars indicate SEM. (E) Quantification by SYBR Green-based qRT-PCR of PBX2 and FRZB mRNA in untreated and Doxy-treated BV173-ShMYB-miR-17-92 cells. Values are reported as 2-ΔCt. GAPDH gene expression was used as endogenous control. Error bars indicate SEM (n=3). (F) (Left panel) Schematic representation of 3’UTRs of FRZB gene with putative binding sites for miR-17-92 cluster. (Right panel) Schematic representation of reporter plasmids containing the wild-type (wt) or mutant (76-81 mut, 1091-1097 mut of miR-17-92-binding sequences) FRZB 3’UTR. Dual Luciferase assay in recipient cells co-transfected with luciferase reporter vectors containing the wt-3’UTR FRZB or the indicated FRZB mutant and either the hsa-miR-17, the hsa-miR-19a mimics or a control (Ctr)-mimic RNA. Firefly luciferase activity of each sample was normalized by Renilla luciferase activity. Results are expressed as fold activation relative to the basal activity of the control mimic (ctr-mimic). (*P≤0.05). The normalized luciferase activity, set as mean of at least three independent experiments performed in duplicate, is shown. Error bars represent the mean±SEM (n=3).