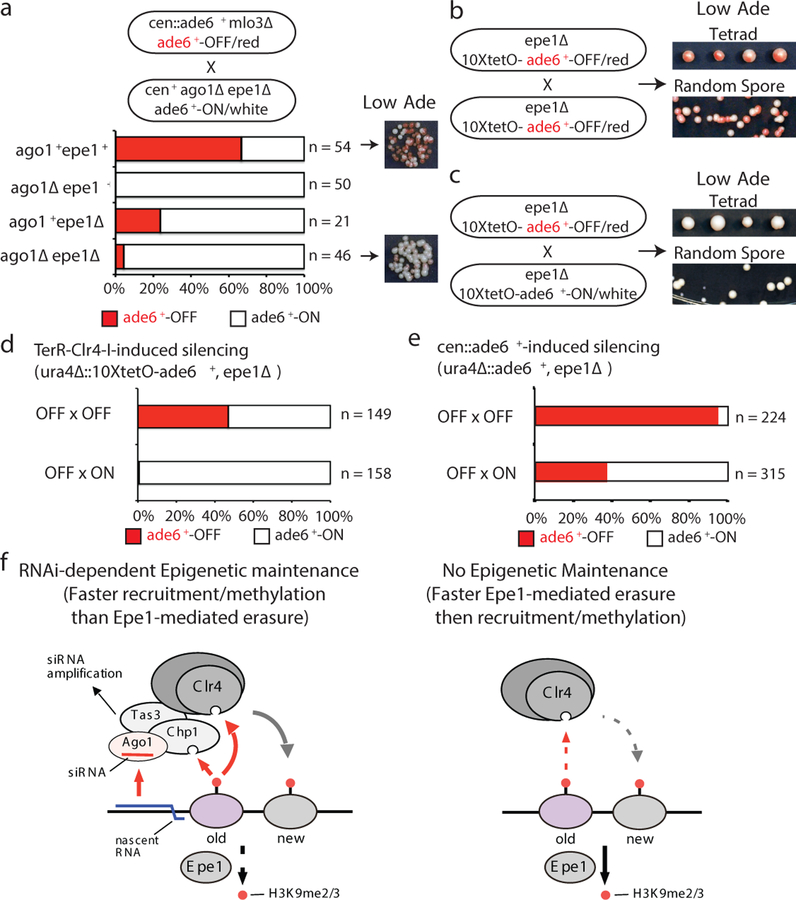
Figure 4 |. RNAi protects an acquired silent state against erasure by Epe1 or activation by an expressed homologous allele.
a, cen::ade6+ mlo3∆ ade6+-OFF cells were crossed to cen+ ago1∆ epe1∆ ade6+-ON cells, followed by random spore analysis. Number and phenotype of mlo3+ progeny with the indicated genotypes and phenotypes are shown. mlo3∆ progeny were excluded. On the right, growth on Low Ade medium indicated meiotic progeny showing maintenance of the ade6+-OFF state in ago1∆ epe11∆ cells. All ago1∆ epe1+ progeny formed white colonies indicating loss of the silent state. n, number of meiotic progeny analyzed.
b-d, epe1∆ ura4∆::10xtetO-ade6+-OFF cells were crossed to either epe1∆ ura4∆::10xtetO-ade6+-OFF (b) or epe1∆ ura4∆::10xtetO-ade6+-ON (c) cells, followed by tetrad dissection (top) or random spore analysis (bottom). Quantification of ON and OFF states in the progeny is shown in d. Silencing was initiated by siRNA-independent TetR-Clr4-I. n, total progeny.
e, Mating of a ura4∆::ade6+-OFF epe1∆ allele with either an OFF or ON allele, followed by diploid formation and sporulation (meiosis), showing that when silencing is established in an siRNA-dependent manner (cen::ade6+ leo1∆), the OFF state is protected from pairing-induced erasure. n, total progeny.
f, Model for the role of RNAi in allele-specific epigenetic inheritance. Left, the siRNA-programmed RITS complex, containing Ago1, Tas3, and Chp1, serves as an epigenetic sensor that maintains allele-specific gene silencing. When both H3K9me and local complementary siRNAs are present, RITS associates with the target locus and recruits the Clr4 methyltransferase complex to methylate histone H3K9 on newly deposited nucleosomes. RITS also promotes local siRNA amplification. Right, in the absence of local RNAi, H3K9me cannot be epigenetically maintained in epe1+ cells.