Extended Data Figure 1 |. siRNA-induced H3K9me3 at the euchromatic ade6+ locus and flanking region.
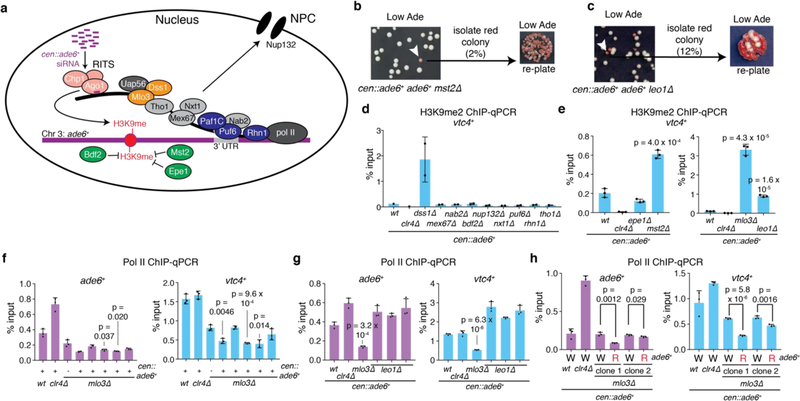
a, Summary of pathways and factors involved in mRNA 3’end processing and its coupling to nuclear export. Chp1 and Ago1 are subunits of the RNAi-induced transcriptional silencing complex (RITS). Uap56 and Mlo3 are TREX (transcription-export) complex subunits, and associate with Dss1 to mediate mRNA export. Tho1 is a subunit of the THO/TREX complex, which is responsible for recruiting other subunits of the TREX complex to mRNA. Mex67 is an ortholog of human NXF1, a critical mRNA export receptor, and associates with Nxt1, another mRNA export factor. Puf6 is an mRNA 3’-UTR-binding protein that has been shown to associate with Mlo3. Rhn1 is involved in RNA Pol II transcriptional termination. Nab2 is a poly(A)-binding protein. The Paf1 complex is required for transcriptional elongation and 3’ end processing, and mutations in Paf1 subunits allow siRNA-mediated heterochromatin formation and silencing of euchromatic genes. Nup132 is a component of the nuclear pore complex (NPC) that has been linked to mRNA export factors. Bdf2 is a histone binding protein that has been shown to inhibit the spreading of centromeric heterochromatin. Epe1 is a jmjC-domain containing putative demethylase that also promotes spreading of heterochromatin. Mst2 is a histone aceyltransferase. See main text for references.
b-c, Around 1000 cen::ade6+ mst2∆ (b) or cen::ade6+ leo1∆ (c) cells were plated on low adenine medium (Low Ade). Most cells formed white colonies, indicating expression of endogenous ade6+, while ~2% of mst2∆ (b) and 12% of leo1∆ (c) cells formed red or pink colonies, indicating silencing of endogenous ade6+ (white arrow). Upon replating, the resulting red colonies formed mostly red colonies, indicating efficient maintenance of the silent state. Repeated once with similar results for each.
d-e, ChIP-qPCR assays showing mean +/− SD H3K9me2 levels at the vtc4+ locus, which is located next to the ade6+ gene, in the indicated mutant cells based on 2 (d) or 3 (e) independent clones. p values are based on a 2-tailed Student’s t-test comparing the indicated mutants to wild-type cells.
f-h, ChIP-qPCR assays showing mean +/− SD Pol II occupancy at the ade6+ (purple) or vtc4+ (blue) locus in mlo3∆ or leo1∆ clones that have not been selected for silencing (99.5% and 88% white, respectively); p values are based on a 2-tailed Student’s t-test comparing the indicated mutants to wildtype cells (f-g). In (h), either an ade6+-ON (W) or ade6+-OFF (R) colony from each of two clones was picked for analysis. p values based on a 2-tailed Student’s t-test comparing the indicated red to white cells for each clone. 3 biological replicates were used per sample.