Abstract
Studies of 3D chromatin organization have suggested that chromosomes are hierarchically organized into large compartments composed of smaller domains called TADs. Recent evidence suggests that compartments are smaller than previously thought and that the transcriptional or chromatin state is responsible for interactions leading to the formation of small compartmental domains in all organisms. In vertebrates CTCF forms loop domains, likely via an extrusion process involving cohesin. CTCF loops interact with compartmental domains to establish the 3D organization of the genome. The continuous extrusion of the chromatin fiber by cohesin may also be responsible for the establishment of enhancer-promoter interactions and stochastic aspects of the transcription process. These observations suggest that the 3D organization of the genome may not be just a determinant, but also a consequence of its function.
Introduction
Chromatin is organized within the three-dimensional (3D) nuclear space, efficiently packaging the genome while allowing proper expression and replication of the genetic material. The relative position of specific loci in the nucleus of individual cells within a population can be visualized using microscopy-based techniques, thus allowing for the understanding of cell-to-cell variation in the arrangement of the chromatin fiber at individual loci. Molecular approaches such as Hi-C can be used to map all interactions between distant loci in the genome, but they require the use of millions of cells and, therefore, provide a view of genome 3D organization that represents an ensemble of the individual cells present in the population1. High-throughput microscopy and single-cell Hi-C are beginning to bridge information obtained using these two approaches2–7.
Here we discuss recent findings suggesting a departure from established dogma in our view of the mechanisms by which 3D chromatin organization is established and its relationship to the regulation of transcription. Recent observations indicate the existence of two independent but partially related organizational principles governing the formation and maintenance of 3D chromatin organization -small compartmental domains that form as a consequence of the transcription/chromatin state, and CTCF/cohesin loops. In the sections below, we first discuss evidence suggesting the existence of compartmental domains, which are established as a consequence of interactions between proteins involved in transcription activation or silencing in each domain. We then analyze evidence in support of cohesin-mediated extrusion as the mechanism underlying the establishment of CTCF loops. Finally, we bring these two sources of information together into a unifying view of 3D genome organization, suggesting that transcription and architecture are closely interdependent and influence each other. Rather than a hierarchical top-down view of nuclear architecture in which 3D chromatin organization determines gene expression, we suggest a balance between compartmental domains and CTCF loops. These observations lead to a new understanding of the causal relationship between transcription and the three-dimensional arrangement of the genome in the nuclear space.
Features of Chromatin Organization
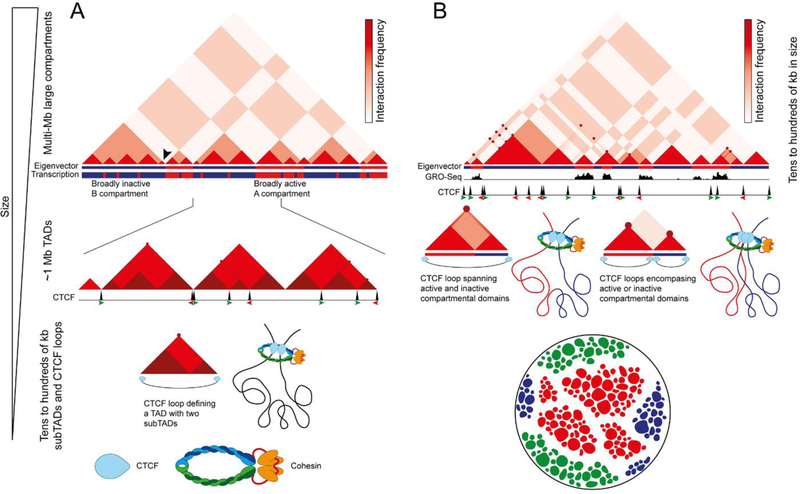
Contact maps of Hi-C data showed a genome-wide view of interactions between all sequences in the mammalian genome for the first time in 20098. These maps displayed a plaid pattern of chromatin interactions over distances as far as the length of a chromosome. These interactions are a manifestation of the segregation of the genome into two compartments, named A and B, defined by the eigenvector or first component of a Principal Component Analysis (PCA)8 (Figure 1A, top). Although the eigenvector gives information on the A/B state, the magnitude of this characteristic, and the region of the linear genome to which this state applies, the published literature generally uses the term compartment to refer to both the domains plus their interactions manifested as the plaid pattern observed in Hi-C heatmaps. Sequences in the A compartment generally contain transcribed genes and active histone modifications, although some regions in A compartments are not transcribed. The same is true for the B compartment, which contains inactive genes with histone modifications associated with a transcriptionally repressed state8 but also some transcribed genes (Figure 1A, top). Due to the high cost of sequencing and the inherent short-range interaction bias of Hi-C, initial maps had low coverage across the two-dimensional genome matrix, with approximately 10 million paired reads18. Thus, sequences were binned in 1 Mb bins for the purpose of identifying compartments using PCA and, consequently, compartments were identified as being multi-Mb in size8. As sequencing costs decreased, Hi-C datasets became richer, with 200–300 million paired reads, allowing the partitioning of data for analysis into ~40 kb bins. Using this smaller bin size (see Box 1), computational algorithms measuring the directionality of interactions in the genome identified TADs as structures in the 0.2–1.0 Mb range9–12 (Figure 1A, middle panel). Whereas A and B compartments correspond to domains that interact preferentially with sequences in other A or B compartment regions, respectively, TADs correspond to sequences that interact preferentially with themselves, rather than with other regions of the genome. TADs are separated by boundaries enriched in CTCF sites and highly transcribed genes. It is important to consider that not all TADs defined computationally at ~40 kb resolution by a directionality index are flanked by CTCF sites9–12 (Figure 1A, middle), although a subset of TADs are defined by CTCF loops (Figure 1A, bottom). The relative sizes of these features has led to a hierarchical model of chromatin organization, in which compartments are composed of several TADs13,14 (Figure 1A, top).
Figure 1. Models of chromatin organization.
A. The hierarchical model of chromatin organization suggests that different sized features contribute to each other’s formation. In this model, compartments are large multi-megabase structures of the 3D genome whereas TADs are substructures inside compartments. Top: Interaction signal (varying intensities of red) from low resolution Hi-C data partitioned into megabase-sized bins show. The panel represents a cartoon version of an actual Hi-C heatmap. The Eigenvector describes the first component of the Principal Component Analysis and identifies A (red) and B (blue) compartments, which correlate with mostly transcriptionally active and inactive regions of the genome, respectively. Middle: TADs are smaller regions of the genome identified with higher resolution Hi-C data partitioned into ~40 kb bins using an algorithm to detect changes in the directionality of interactions. The panel shows a small section of the genome corresponding to one B compartment and half A compartments in the diagram above. TATs contain smaller subTADs characterized by higher interaction frequencies (darker shade of red) and CTCF loops detected as strong punctate signal corresponding to strong interactions between CTCF sites. Note that only some TADs coincide with CTCF loops and CTCF is only present at the borders of some TADs. Only some CTCF loops are detected at this resolution. Bottom: Structure of a TAD as detected at ~40 kb resolution, containing two subTADs and flanked by CTCF/cohesin sites forming a loop.
B. An alternative model of chromatin organization incorporates recent findings obtained with very high-resolution data partitioned in 1–5 kb bins. Top: The cartoon corresponds to the domain marked with an arrowhead in panel A and it is a representation of the actual Hi-C heatmap, emphasizing the complexity of interactions present in a region that appears as a uniform minute TAD in low resolution data. The Eigen vector obtained by binning the data at 5–20 kb allows the identification of compartmental domains, which accurately correspond to the active or inactive transcriptional state determined by GRO-seq. Punctate signal represent CTCF loops between sites in convergent orientation. Middle: Some CTCF loops encompass active and inactive compartmental domains, increasing interactions between these two domains that would normally not take place (left). Other CTCF loops encompass individual compartmental domains, and the formation of the loop decreases interactions between two adjacent domains (right). Therefore, the presence of CTCF loops modulates interactions among compartmental domains. Bottom: Segregation of chromatin states in the nucleus may occur as a consequence of the presence of different classes of multivalent proteins that mediate class-specific interactions to create different phases, wich result in droplets of distinct chromatin states within the nucleus. In the cartoon, red represents proteins and histone modifications present at genes or regulatory sequences in a transcriptionally active state, blue represents H3K27me3 and Polycom-Group proteins, and green represents H3K9me3 and HP1.
The first high-resolution Hi-C dataset of a mammalian genome contained ~5 billion paired reads, making it possible to bin reads at 5 kb resolution. The use of the Arrowhead algorithm allowed the identification of contact domains, smaller in size than TADs. A subset of these contact domains arise as a consequence of point-to-point interactions between two sequences bound by CTCF. These loops are relatively stable, since they can be observed in high resolution Hi-C data as a strong punctate signal at the summits of some domains19 (Figure 1B, middle). Given the population nature of Hi-C data, these strong spots can be interpreted as interactions occurring in all the cells of the population or as very stable interactions taking place in a sub-population of cells. In addition to these CTCF loop domains, a second type of contact domains, originally named “ordinary domains”, are characterized by the presence of specific histone modifications and are not flanked by CTCF19 (Figure 1B, middle). Identification of compartments using PCA and high-resolution Hi-C data binned at 10–50 kb suggests that compartments are smaller than previously thought, as small as a single active or inactive locus20 (Figure 1B, middle; compare eigenvector with domains above represented by red triangles). Therefore, the ordinary domains likely correspond to small compartments formed by the segregation of active and inactive chromatin19,20. In the rest of the manuscript, we will use the term “compartmental domains” to refer to domains identified by PCA using high resolution Hi-C data and bin sizes in the range of a few kb, whereas we will use the term compartments to refer to the original domains identified using bins in the hundreds of kb to 1 Mb range. Compartmental domains, as is the case for compartments, can be classified into active A and inactive B domains, are smaller than TADs, and are present inside, between, or overlapping CTCF loops in mammals20,21 (Figure 1B, middle). CTCF loops can sometimes encompass two compartmental domains with different transcriptional states, increasing interactions between them (Figure 1B, bottom left). In other cases, CTCF loops only contain sequences in the same transcriptional state and decrease interactions between two adjacent compartmental domains (Figure 1B, bottom right). These observations suggest that, instead of being composed of hierarchically-related large compartments and smaller TADs, chromosomes are organized into similarly sized compartmental domains and CTCF loops. High resolution Hi-C maps also allowed the identification of compartmental domains in Drosophila melanogaster, where A compartmental domains have a median size of just 15 kb20. Their size corresponds to the median size of transcriptionally active blocks of chromatin, a finding that suggests a relationship between compartmental domain formation and chromatin state20. Compartmental domains are also found in many other organisms such as Arabidopsis thaliana, Oryza satvia, Zea mays, Caenorhabditis elegans, Neurospora crassa, and Plasmodium falciparum, and likely explain most aspects of 3D chromatin organization in these organisms20,22.
Compartmental Domains and Transcription
The close correlation between A and B compartmental domains and the transcriptionally active or inactive state of chromatin suggests a possible causal relationship between the two. Indeed the correspondence between chromatin organization and transcriptional state is sufficiently precise to accurately predict Hi-C maps in many different organisms using global run-on sequencing (GRO-seq), RNA sequencing (RNA-seq), or histone modifications20,23–25. When discussing evidence supporting this relationship, we will distinguish between transcription per se versus the transcriptional state i.e. proteins or histone modifications normally associated with expressed or silenced genes, even if the genes in a transcriptionally active state are not actually transcribed. Several studies have tried to establish a possible role for transcription in chromosome organization using chemical inhibition of the initiation or elongation of RNA polymerase II (RNAPII). Some drugs used to inhibit transcription also affect RNAPII levels at the promoter whereas others do not. Therefore, when interpreting results from this type of experiments, it is important to consider whether only transcription has been affected or whether the presence of proteins in the transcription complex, which may be responsible for mediating interactions between A compartmental domains, has been affected too. In the prokaryotes Caulobacter crescentus and Bacillus subtilis, inhibition of transcription using rifampicin results in a loss of contact domains26,27. However, similar experiments in eukaryotes have led to more nuanced observations. Inhibition of transcription initiation in D. melanogaster cells using triptolide results in a reduction of interactions inside compartmental domains as well as a reduction in the plaid pattern representing contacts between compartmental domains20,28. In contrast to prokaryotes, inhibition of transcription in D. melanogaster does not eliminate compartmental domains entirely. One possible explanation is that the maintenance of these domains is dependent on the presence of proteins related to transcription, rather than the transcription process. Under the conditions used to inhibit transcription with triptolide, a significant amount of RNAPII, and perhaps other components of the transcription complex, remain at the promoter20. Indeed, the degree of RNAPII loss after treatment correlates with the degree to which compartmental domains are reduced20. Furthermore, the D. melanogaster heat shock response, which results in the downregulation of most genes, has a stronger effect on RNAPII levels than triptolide treatment and causes a more pronounced loss of compartmental domains20,28.
Studies of the relationship between transcription and the formation of compartmental domains have also taken advantage of the gradual establishment of normal transcription during early embryonic development. In D. melanogaster, embryos undergo 12 nuclear divisions before global transcription can be detected by standard methods. During nuclear cycle 12 (nc12), a few genes are transcribed but most are inactive. These few transcribed genes correspond to the few compartmental domains present at this stage29. After genome-wide transcriptional activation during nc13 and nc14, new domains form around the newly transcribed loci29. These observations support a relationship between transcriptional activation and compartmental domain formation during D. melanogaster development. To test this relationship, alpha-amanitin or triptolide were used to inhibit transcription at an early embryonic stage in order to prevent normal genome-wide transcriptional activation. This inhibition resulted in decreased compartmental domain formation, suggesting that transcription may be responsible for organizing chromatin into domains during D. melanogaster embryonic development29. However, compartmental domains were not entirely eliminated, and their intensity correlates with the amount of RNAPII left at gene promoters after transcription inhibition20. These results support the conclusion that proteins related to the transcriptional state, rather than transcription itself, may be responsible for the establishment of compartmental domains.
Similar experiments have been carried out with mammalian embryos, which start transcribing at the two-cell stage. Results from Hi-C experiments performed with mouse embryos at different stages of embryonic development suggest that the appearance of contact domains correlates with the time of transcription activation, with weak interactions observed at the 2-cell stage that strengthen in the 8-cell embryo31,32. Treatment with alpha-amanitin starting at the zygotic pronuclear 4 (PN4) stage and continuing for two days, a time in which embryos would normally mature to the 8-cell stage, instead results in embryos arrested at the 2-cell stage32. Hi-C maps of these arrested embryos show slightly stronger domains than those present in untreated 2-cell stage embryos, but weaker domains than the normal 8-cell stage. When compared to normal 8-cell stage embryos, the results suggest an effect of transcription inhibition on compartmental domain organization. However, since the treated cells are arrested at the 2-cell stage, but their age is that of control 8-cell embryos, this result is difficult to interpret as it is unclear to which stage they should be compared. In a similar study, transcription was inhibited using alpha-amanitin beginning at PN3 and lasting for 20 or 45 hours, when embryos would normally proceed to late 2-cell or 8-cell stages, respectively. In both cases, embryos were arrested at the 2-cell stage31. Hi-C maps of these embryos show that, while transcriptional inhibition did not completely stop the progression of chromatin organization during embryonic development, many features of chromatin organization do appear less pronounced (see Du et al. 2017 Extended data 8b and compare Extended Data 8d with Extended Data 4e)31. These results may be interpreted to suggest that transcription may not play a large role in mammalian chromatin organization. However, both CTCF loops and compartmental domains contribute to the establishment of 3D architecture, and inhibiting transcription may only affect compartmental domains. Thus the presumably unchanged CTCF loops after transcriptional inhibition may not allow the detection of changes in compartmental domains due to the relatively low Hi-C sequencing depth used in these experiments31,32. Although technically difficult, the definitive answer to these questions will require an analysis of the distribution of CTCF and cohesin during early mammalian embryogenesis, as well as the effect of transcription inhibitors on the actual transcription rate and the levels of RNAPII and other transcription factors at the promoter of genes. This will distinguish between a requirement for transcription per se versus a requirement for factors in the transcription complex that mediate interactions leading to the formation of compartmental domains.
Additional insights into the relationship between the formation of compartmental domains and transcription come from the analysis of 3D chromatin organization in the mature oocyte and sperm, the two cells whose genomes will contribute to the one-cell zygote. The mature oocyte is arrested in metaphase II of meiosis and its genome is not transcribed. As expected from previous analyses of 3D chromatin organization in mitotic chromosomes, Hi-C maps reveal an absence of long-range intra-chromosomal interactions in the nucleus of mature oocytes31–33. However, although also transcriptionally inactive, sperm chromosomes are organized in the 3D space in a manner similar to that of embryonic stem or somatic cells, with clear compartmental domains and CTCF loops31,32,34,35. One explanation for this observation is that 3D organization is established in round spermatids, which are transcriptionally active, and it is maintained in sperm. Alternatively, sperm retain transcription factors and nucleosomes with specific histone modifications at a subset of particular sites, including transcription start sites (TSSs) and distal intergenic sites presumed to be regulatory sequences34. It is possible that proteins present at these sites mediate interactions that result in the formation of compartmental domains. It is unclear whether these domains are maintained in the zygote or whether they disappear as protamines present in the sperm are replaced by nucleosomes and then re-established as new transcription is initiated in 2-cell embryos.
Formation of Compartmental Domains
Compartmental domains in D. melanogaster and mammals are composed of one or more adjacent genes in the same transcriptional or chromatin state20,21. These domains are the result of interactions among sequences located within the domain. Most frequent among these interactions are those taking place between the start and termination sites of transcribed genes20. These interactions cause the formation of gene loops, resulting in a large accumulation of proteins involved in transcription at a single site, including transcription factors bound to enhancers, the transcription complex at the promoter, and proteins involved in splicing and transcription termination. In D. melanogaster, approximately 15 different architectural proteins, including CTCF, may also contribute to this local increase in protein concentration20,36. These proteins, which were originally identified based on their insulating effects on enhancer-promoter interactions, do not form stable loops as CTCF does in vertebrates20,37. Instead, D. melanogaster architectural proteins and associated RNAs bind to genomic sites containing multiple DNA binding motifs and located in close proximity to promoters, perhaps contributing to an increase in the local protein concentration at these sites. Interestingly, some of these proteins are modified by sumoylation or parylation, which could amplify their ability to interact with other proteins able to bind SUMO or poly(ADP-ribose) (PAR)36. It is therefore tempting to speculate that insulator bodies, which can be visualized in the nucleus as nuclear bodies or membraneless organelles with antibodies to any of the D. melanogaster architectural proteins, are the result of inter-compartmental domain contacts mediated by cooperative interactions among architectural and other proteins involved in the transcription process37–40.
The plaid pattern observed in Hi-C heatmaps from a population of cells is likely a combination of individual interaction patterns present in each cell of the population. Inter- compartmental domain interactions are stochastic and their frequency or stability may depend on the number, affinity, and interaction ability of the proteins involved, which determine the cooperativity of the interactions38. It has been recently proposed that cooperative interactions among large numbers of multivalent transcription factors can be explained by a phase separation model38,39. Based on this model, active and inactive regions of the genome - A and B compartmental domains - containing two different sets of multivalent proteins, respectively, may be able to interact with members of their own class, forming two different phases that preclude inter A-B compartmental contacts. For example, it was recently shown that phase-separated heterochromatin protein 1α (HP1α)-mediated heterochromatin droplets are formed in vitro and can be detected in vivo40. Similarly, phase-separated droplets of active chromatin may be possible by proteins that contain intrinsically disordered domains, such as those commonly found in transcription factors and RNAPII41,42. Phase separation of chromatin into droplets could regulate functional aspects of compartmental domain interactions. For example, droplet formation may increase the concentration of transcription factors and RNAPII at active chromatin, analogous to the transcription factory hypothesis43. A model of phase-separated chromatin (Figure 1B, top) would entail the constant fusion and fission of chromatin droplets suggesting that compartmental domains are involved in dynamic interactions. The dynamics of such droplet activity in the cellular population could explain why active compartmental domains appear to interact with every other active locus across the length of the chromosome in Hi-C heatmaps8,19.
CTCF Loop Domains can be Explained by Extrusion
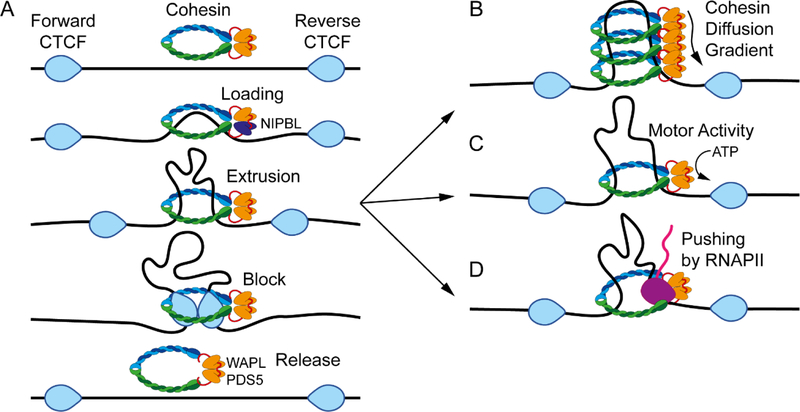
In addition to compartmental domains and their associated long-range intra- chromosomal interactions, high resolution Hi-C maps of mammalian cells show thousands of intense, highly localized, punctate signals that correspond to loops anchored at CTCF sites19 (Figure 1B, middle). Ninety two percent of CTCF loops identified by Hi-C, or 65% identified by CTCF ChIA-PET, occur between motifs in a convergent forward-reverse orientation19,44 (Figure 2A). Most other CTCF loops are formed between motifs oriented in forward-forward or reverse- reverse orientations, but are comparatively weak in Hi-C experiments, making them more easily detectable by CTCF ChIA-PET19,44. Therefore, both Hi-C and CTCF ChIA-PET show that CTCF loops preferentially occur between motifs in convergent orientation, with a strong bias against the opposite orientation. The importance of this orientation preference has been demonstrated by CRISPR-mediated inversion of CTCF motifs at individual loci, which alters the corresponding loop domain and allows the formation of new enhancer-promoter interactions45. The consistency in the preference for convergent CTCF motifs at loop anchors is also underscored by the ability to accurately predict changes in CTCF loops after CRISPR-mediated inversion or deletion of CTCF motifs46. These findings have important implications to explain how CTCF bound at a specific site finds its partner site to form a loop. If DNA-bound CTCF is able to diffuse unrestricted in the 3D space to encounter the second loop anchor at tens or hundreds of kb away, the orientation of the CTCF-bound motif should be irrelevant to the establishment of CTCF-CTCF interactions and the formation of a loop. Experimental observations indicating a preference for motifs in a convergent orientation serving as loop anchors suggest that CTCF molecules encounter each other in a dimension-restricted space. This can be accounted for if the formation of CTCF loops takes place via an extrusion process mediated by the cohesin ring46–50.
Figure 2. Mechanisms of loop extrusion.
A. General model of loop extrusion. The extrusion process involves cohesin composed of SMC1, SMC3, and RAD21 which is loaded onto chromatin via NIPBL. Extrusion is blocked at CTCF sites arranged in a convergent head to head orientation. Some proportion of cohesin is released throughout this process by the activity of WAPL and PDS5.
B. Extrusion via cohesin diffusion. Extrusion may occur by constant loading of cohesin resulting in a diffusion gradient.
C. Extrusion via cohesin motor activity. An alternative explanation for extrusion is that the process is driven by the motor activity of cohesin via ATP hydrolysis.
D. Extrusion via pushing of cohesin by RNAPII. Other factors able to move along chromatin, such as RNAPII (purple), may help cohesin to extrude DNA.
The loop extrusion model suggests that structural maintenance of chromosomes (SMC) proteins, as part of cohesin or condensin, progressively extrude chromatin until blocked by CTCF bound to a properly oriented site46–50 (Figure 2A). Several groups have investigated the mechanics of SMC-mediated loop formation, and multiple observations suggest that cohesin and condensin rings can topologically entrap and move along the DNA until meeting an obstacle that blocks this movement. For example, in vitro experiments have shown that cohesin can diffuse along anchored DNA, a process that can be blocked by CTCF51–53. It was also shown that condensin can translocate a second piece of DNA relative to the first and, more recently, Ganji et al were able to visualize condensin mediated loop extrusion in vitro54,55. Condensin mediated extrusion of naked DNA can occur via a single ring, but it is not known whether this also happens with chromatin templates or whether a single cohesin ring can also extrude DNA in vitro54. One cohesin ring can capture two separate pieces of DNA, but only if the second piece of DNA is single stranded56. This provides some evidence of cohesin mediated extrusion through a single ring, but more work is needed to fully understand the process on a chromatin template in vivo. In testing the extrusion model, polymer physics simulations have suggested that cohesin starts randomly in the genome and the extrusion process is continuous until blocked by CTCF when approaching from the 3’ end of the motif i.e. inside the loop47 (Figure 2A). In support of this, results from ChIP-exo and ChIP-nexus experiments show that cohesin is enriched at the 3’ end of CTCF motifs44,57. Additionally, while lack of cohesin results in loss of CTCF loop domains21,58,59, depletion of the cohesin release factor WAPL causes cohesin to remain bound to chromatin for longer periods, resulting in the formation of larger CTCF loops59–61. This suggests a processive mechanism of loop formation, such that the size of CTCF loops corresponds to the amount of time that cohesin is able to extrude chromatin before encountering an obstacle that stops extrusion.
How CTCF provides a barrier to extrusion is unknown, but it may be related to CTCF induced conformational changes to chromatin. CTCF binding repositions nucleosomes62 and was recently shown to cause large changes to naked DNA in vitro63. Experiments using atomic force microscopy showed that DNA wraps around the bound CTCF protein forming CTCF centric circles ~67–80 nm in diameter63. These CTCF-DNA circles are larger than the ~20 nm proteins that are able to block cohesin sliding in vitro51,52. However, this fails to explain the unidirectional blockage of extrusion coincident with CTCF motif orientation or the fact that the presence of CTCF and cohesin alone cannot fully explain the formation of loops between specific sites in the genome, since many CTCF peaks detected by ChIP-seq do not form loops even when in convergent orientation19,64,65. Additionally, many loops appear to change during differentiation, often without changes in CTCF binding66,67. For example, experiments using Hi-C resulted in the identification of 184 loops gained and 33 loops lost during macrophage differentiation without alterations in CTCF occupancy66. Interestingly, these loops are enriched in the AP-1 motif, suggesting that transcription factors may play a role in regulating loop formation between CTCF bound loci66. Thus, although much of the focus on the establishment of CTCF loops has centered on this protein, future work should also focus on the possible role of transcription factors in CTCF loop formation.
Mechanisms of Loop Extrusion
The mechanistic details of the process by which loops are extruded by the cohesin ring are now beginning to be understood. Analysis of 3D chromatin organization of metaphase chromosomes suggests that all compartmental domains, their interactions, and CTCF-mediated loops disappear as condensin-mediated loops progress to condense chromosomes during mitosis33,68. This indicates that cells need to reconstruct the 3D organization of their genomes when they exit mitosis. Since compartmental domains appear to be a consequence of the transcriptional or chromatin state of the genome, the activation of transcription at the M/G1 boundary should be sufficient to restore this aspect of 3D organization after the loss of condensin mediated chromosome condensation. Alternatively, the memory of chromatin state via histone modifications or other proteins that remain bound during mitosis may be sufficient to restore compartmental domains in the absence of actual transcription. However, re- establishment of CTCF loops will require cohesin-mediated extrusion, which presumably will necessitate large amounts of ATP and will have to be done rapidly. Recent experiments examining the re-establishment of CTCF loops after cohesin removal have shed light on the issue of how rapidly cohesin can extrude to restore CTCF loop domains. Lieberman Aiden and colleagues used auxin-mediated degradation of RAD21 to examine this process. After removal of RAD21 for a 6 h period, all CTCF loops are eliminated, indicating that CTCF cannot stabilize loops without cohesin involvement. This supports a model whereby loops and the associated domains are dynamic features that may require constant extrusion47,69. Restoring cohesin by removal of auxin results in the re-establishment of CTCF loops as large as 900 kb within 40 minutes21. If loop extrusion were to use one cohesin complex per loop, this suggests that the speed of extrusion by cohesin is at least 375 bp/s or that each of the two topologically entrapped sections of DNA are pulled through cohesin rings at 188 bp/s. This is likely a conservative estimate, since it does not account for the time required for cohesin to re- accumulate in the nucleus and to be loaded onto chromatin before extrusion can begin again. Analysis of chromosome organization in B. subtilis using Hi-C at several different time points also gives insights into the speed of the extrusion process. These experiments allowed the visualization of the progressive “zip-up” of DNA from the origin by an SMC protein complex26,70. The estimate of the rate of extrusion in bacteria based on these experiments is approximately 850 bp/s26. A second study using real-time imaging of loop extrusion in vitro found that condensin extrudes at ~600 bp per second54. Thus, while the actual extrusion rate in mammals is unknown, it is likely somewhere between 374 and 850 bp/s, suggesting a sufficiently fast process to account for the need to rapidly form loops at different stages of the cell cycle.
The mechanisms by which cohesin can translocate along the DNA at such speeds are not known, but several models have been put forward to explain this phenomenon, including diffusion, motor activity, and pushing by other macromolecular assemblies. Diffusion of cohesin by Brownian motion along the 10 nm chromatin fiber may explain the forces controlling the extrusion process (Figure 2B). Simulations of this model have suggested that diffusion may reach high speeds depending on the concentration of cohesin loaded71. In agreement with this model, analysis of loops re-established after cohesin removal and restoration shows that those containing more of the cohesin loader NIPBL recover sooner than loops containing lower levels of this protein21. It is tempting to speculate that the greater amount of cohesin loaded on chromatin results in an increased diffusion rate and extrusion of the loops. However, the correlation between recovery time and the number of NIPBL binding sites may also be explained by a higher probability of cohesin loading at an early stage in the recovery period, or NIPBL may enhance loop enlargement in other ways. Thus, whether cohesin diffusion can explain loop extrusion without additional help is unclear and requires additional experimental evidence.
SMC complexes possess ATPase activity, and it is possible that they act as their own motors to translocate chromatin using energy from ATP (Figure 2C). The condensin complex was shown to move in vitro in a randomly chosen but single direction55. This unidirectional movement requires the ATPase domain, indicating that energy consumption is necessary for motor activity55. It was calculated that condensin moves 30 bp per ATP hydrolyzed55. The average CTCF loop size in humans is 180 kb19, which would require approximately 6000 molecules of ATP per loop, assuming that in vitro studies are an accurate representation of in vivo function. When considering an upper estimate of 50,000 CTCF loops in the genome19,44, forming all of them once would require an average of approximately 3X108 molecules of ATP. This energy requirement is more or less in line with other energy estimates suggesting the need for 4×105 molecules of ATP per second for the movement of a cell or 2×107 molecules of ATP per second for the duplication of its proteome once every 24 hours72. Therefore, the energy requirement to form CTCF loops by extrusion powered by the cohesin ATPase activity is in line with other energy requirements of the cell. However, the estimate of energy needed would increase considerably when considering the idea that loops are continuously extruding and/or have multiple extrusion complexes per loop47. It should also be noted that these measurements of motor activity were performed in vitro on naked DNA using the condensin complex from yeast, and it is possible that mammalian cohesin may have different rates of processing in vivo. Indeed there is evidence that proteins bound to DNA slow down or block cohesin movement in vitro51,52. Determining whether cohesin acts as its own motor and if the cell devotes the required energy for loop extrusion is a critical issue supported by recent results suggesting that ATP is required for CTCF loop formation73. In these experiments, cells were initially depleted of cohesin to eliminate CTCF loops, then cohesin was allowed to recover under normal conditions or after depletion of ATP. Cells under normal conditions were able to re-establish CTCF loops, whereas those depleted of ATP were not, indicating that CTCF loop formation requires ATP, probably during the extrusion process73.
Cohesin may also be pushed along chromatin by unknown translocating factors, one of which may be RNAPII (Figure 2D). Movement of cohesin as a consequence of the transcription process has been shown in vitro51,52. Also, induction of transcription in vivo relocates cohesin to the 3’ end of convergently oriented genes in yeast52,74,75. In bacteria, transcription affects progress of the SMC complex as it zips up the chromosome26,70. Changing the orientation of transcription to be contrary to the zip up direction antagonizes the progress of SMC and influences interactions26,70. Thus, RNAPII may push the SMC complex and thereby influence chromatin organization. In mammals, cohesin is also thought to be pushed by transcription52,76. Deletion of the cohesin release factor WAPL in combination with deletion of CTCF results in cohesin relocation to the 3’ end of genes, similar to what was shown in yeast76. Cohesin relocation by transcription could be due to direct pushing by RNAPII74 or, indirectly, by chromatin supercoiling77. In support of this, it was recently found that inhibition of transcription elongation by flavopiridol results in a moderate decrease in CTCF looping, though this effect is not as strong as that of ATP depletion73. In spite of this evidence, loop extrusion via RNAPII thrusting fails to explain how loops form in inactive regions of the genome. Additionally, the speed of loop extrusion, which is at least 374 bp/s as discussed above, does not fit with current estimates that place RNAPII elongation rates at 9 to 90 bp/s21,78. These issues question the feasibility of transcription as the driver of loop extrusion. Alternatively, instead of pushing cohesin, the slow elongation rate may suggest that RNA polymerase may interfere with cohesin movement, and thereby slow down the extrusion process over transcriptionally active regions.
Although it is unclear which of these models best describes loop extrusion (Figure 2), analyses directed towards understanding the translocation speed, energy consumption, and relationship between transcription and SMC complex movement will be informative in deciding among them. While there is some evidence supporting each model, each has its own limitations, and it is possible that a combination of these mechanisms underlies the extrusion process. This process, if continuous and random, gives rise to CTCF loops at locations where extrusion is stopped by this protein while bringing together sequences located within the same or different compartmental domains. Therefore, forces underlying the extrusion process must coexist with those responsible for interactions among compartmental domains in the same transcriptional state. Therefore, these two types of domains must influence each other, a matter that we discuss next.
CTCF loops versus Compartmental Domains in 3D organization
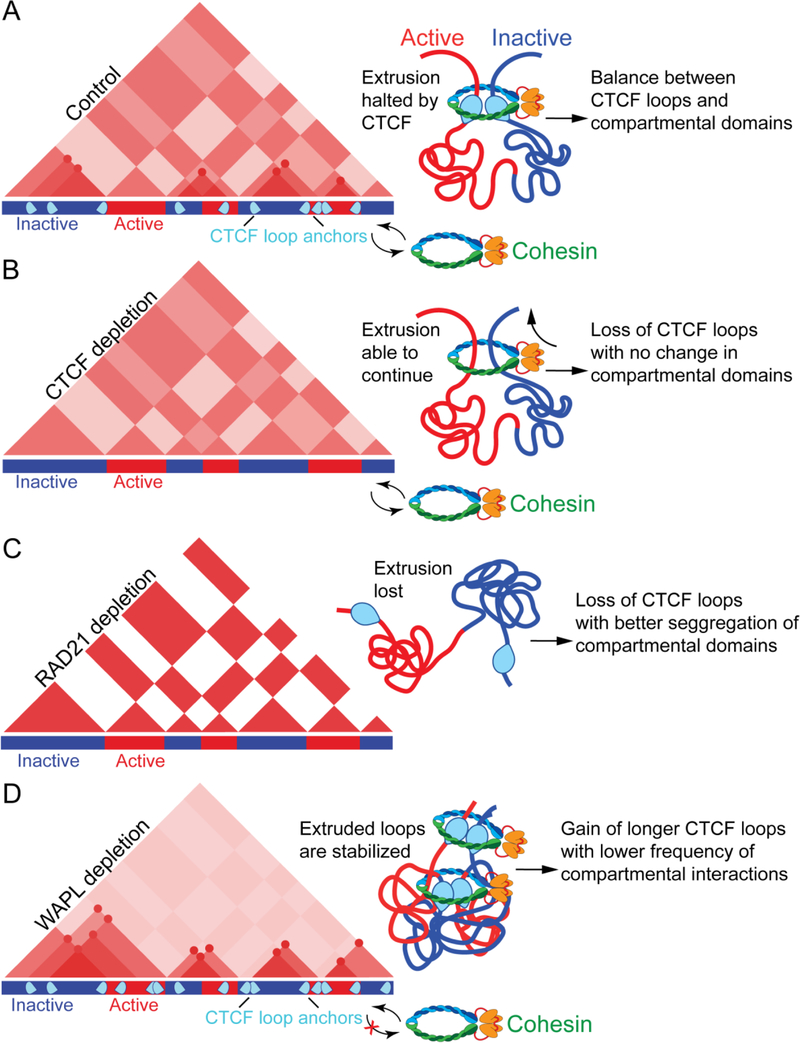
Complete depletion of CTCF results in embryonic lethality in mice79, but the use of auxin-mediated degradation approaches has allowed several recent studies to examine its role in 3D chromatin organization. After depletion, CTCF loops disappear, yet interactions among and within compartmental domains remain59,64,65 (Figure 3). Indeed, CTCF loops and compartmental domains seem to form independent of each other, supporting a model where chromosomes are organized by two distinct but partially inter-dependent features, compartmental domains and CTCF loops.
Figure 3. Effects of CTCF, cohesin, or WAPL depletion on 3D chromatin organization.
A. Chromatin is organized in the 3D nuclear space by CTCF loops and compartmental domains. Some CTCF loops restrict the ability of active (red) and inactive (blue) regions to segregate into compartmental domains whereas others increase the frequency of interactions between two adjacent active and inactive domains (top right).
B. Depletion of CTCF results in a loss of CTCF loops but no change in compartmental domain interactions, likely because cohesin is able to randomly continue extruding chromatin.
C. Depletion of cohesin results in loss of CTCF loops, more distinct compartmental domains and stronger inter-compartmental interactions. D. Depletion of WAPL results in gain of longer CTCF loops and decrease of interactions among compartmental domains.
Like CTCF, cohesin is also important for loop formation. A conditional deletion of Nipbl, the cohesin loader (see Figure 2A), was recently used to prevent cohesin from loading onto chromatin58,60. This deletion does not affect CTCF occupancy, but results in widespread loss of CTCF loops. Unlike CTCF, Nipbl deletion results in stronger segregation between A and B compartmental domains58. In similar studies, depletion of the RAD21 subunit of cohesin (see Figure 2A) or deletion of other subunits causes a widespread loss of CTCF loops21,59,61. Unlike CTCF depletion, but similar to the loss of Nipbl, the depletion of RAD21 results in increased segregation of active and inactive regions into compartmental domains21,59. This is manifested by more defined squares in the Hi-C checkerboard pattern (Figure 3). The different results obtained after depletion of CTCF or cohesin suggest that cohesin mediates interactions other than those involved in CTCF-CTCF contacts. In CTCF depleted cells, cohesin is likely able to extrude randomly and thereby may prevent complete segregation of compartmental domains. Indeed, inducible deletion of CTCF results in a widespread relocation of cohesin away from CTCF motifs76. A few recent studies also examined what happens to chromatin organization when cohesin is blocked from disassociating from chromatin by depleting the cohesin release factors WAPL or PDS5 (see Figure 2A). Loss of either of these two proteins cause no dramatic change to existing CTCF loops, but new loops are formed spanning larger distances than those present in wild-type cells59–61. This suggests that the residency time of cohesin determines loop size. Thus, extrusion may be only partially blocked, or perhaps stalled, by each convergently oriented CTCF site, and may continue extruding until released by WAPL or PDS5. Interestingly, the increased residency time of cohesin after WAPL or PDS5 depletion results in an increase in short-range contacts and decreased long-range interactions between compartmental domains59,60 (Figure 3). Without CTCF, the length and genomic location of extruded regions will depend upon the relative ratio of loading and unloading by NIPBL and WAPL, respectively. Loading and unloading of cohesin are thought to occur randomly, which would cause the formation of random loops in each individual cell. Because there is no precise extrusion stopping point without CTCF, these loops would appear as random signal when averaged across the cell population. When NIPBL or RAD21 are depleted, the loss of extrusion-mediated local interactions could allow compartmental interactions to more easily occur over long distances. Inversely, when WAPL or PDS5 are depleted, increased local interactions as a result of continuous extrusion may reduce the ability of long-range inter-compartmental domain interactions to take place (Figure 3).
CTCF Loops versus Compartmental Domains in Gene Regulation
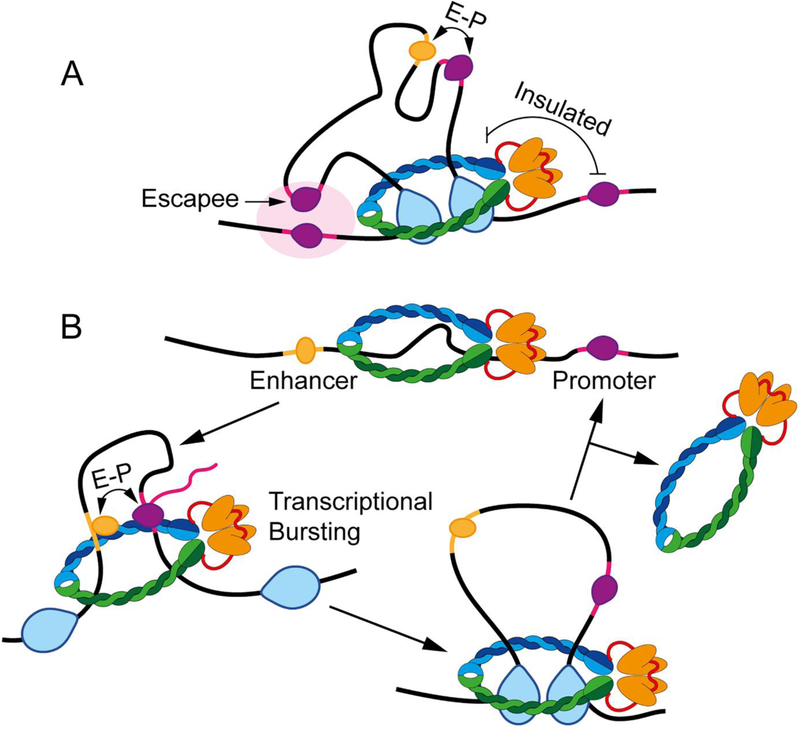
Previous studies indicate that CTCF and cohesin play two functionally distinct roles: to help enhancers find their cognate promoters and to restrict enhancer-promoter interactions between sequences located inside and outside CTCF loops15,80. These two apparently opposite functions of CTCF can be explained based on the requirement of extrusion to form CTCF loops. CTCF represents a barrier to loop extrusion, and the formation of a loop domain results in increased interactions within the loop (Figure 4A). The involvement of cohesin in the extrusion process explains its requirement for interactions not only between the anchors but also within loop domains21,58–60. Therefore, enhancers and promoters that lie in the interior of the loop may interact more frequently with each other than in the absence of a loop. The extrusion process also explains why enhancers and promoters that lie on either side of a loop anchor are less likely to interact (Figure 4A). However, the extrusion process that results in the formation of CTCF loops does not completely preclude interactions between sequences on either side of a loop anchor. Active compartmental domains located inside a loop can partially escape CTCF loops and interact with sequences in other active domains located outside of the CTCF loop20 (Figure 4A). Importantly, because cohesin may move past CTCF anchors at a low frequency69, this could explain why long range interactions between compartmental domains are able to escape the constrains of CTCF loops.
Figure 4. CTCF loops and enhancer-promoter interactions.
A. CTCF loops establish domains in which sequences can interact more frequently. These contacts are thought to help promote enhancer (yellow with orange transcription factor) – promoter (pink with purple RNAPII) (E-P) interactions when inside the domain, but help insulate against those outside the domain. However, examples of genes that escape the CTCF domain and interact with adjacent sequences can be observed in Hi-C data (arrow). It is likely that these “escapee” genes interact with promoters or regulatory sequences within A compartmental domains (large light pink oval).
B. A speculative model of transcriptional activation. In this model, genes are inactive when extrusion has not begun (top) and are activated once extrusion brings together enhancers and promoters (left). Gene activity is lost once extrusion moves past the enhancer or promoter (right), but will be reestablished during each extrusion event. Regular extrusion events causing gene activation at discrete times may explain transcriptional bursting.
These observations suggest a critical role for CTCF in gene expression. However, the effects of CTCF removal on transcription can be quite variable, depending on the situation. Complete loss of CTCF is lethal during embryonic development while haploinsufficiency results in intellectual disability, microcephaly, and growth retardation79,81,82. Heterozygous CTCF- knockout mice show a high incidence of tumors, and mutation of specific CTCF binding sites correlates with various cancers in humans83,84. Accordingly, changes in CTCF looping at specific genomic sites have effects on the expression of nearby genes. For example, deletion of CTCF motifs at the HoxA locus results in increased interactions between active regions and genes that are normally repressed in motor neurons. In agreement with a model where the formation of CTCF loop domains increases interactions between enhancers and promoters, this increase in interactions corresponds to a large increase in gene expression85. Similarly, CRISPR-mediated inversion of individual CTCF sites at the Pcdh-α locus results in the loss of interactions between the HS5–1 enhancer and Pcdh-α promoters, with a corresponding decrease in gene expression. This inversion also results in a gain of interactions between the HS5–1 enhancer and genes in the Pcdh-β locus. However, instead of a gain in gene expression corresponding to the increased interactions with an active enhancer, Pcdh-β genes display decreased gene expression45. In a different study, CRISPR inversion of individual CTCF sites resulted in changes in interactions but mild changes (~1.5–2.5 fold) in gene expression86. In the context of these findings, it is surprising that general depletion of CTCF or cohesin in various cell types under cultured conditions has very small immediate consequences on transcription, although cells depleted of CTCF die after 4 days in culture64. For example, knockdown of CTCF in HEK293T cells results in only 161 differentially expressed genes87. This small effect may have been influenced by the residual amount of CTCF remaining after knockdown. However, nearly complete depletion of CTCF via auxin degradation only results in 370 differentially expressed genes, of which only 43 show at least a 5-fold change in expression64. A separate study in which CTCF loops were eliminated by RAD21 degradation found only 2 genes with at least a 5-fold change in gene expression21. These findings suggest that gene expression is often surprisingly resilient to acute changes in CTCF loops or cohesin-mediated extrusion. Reconciling the drastic consequences of CTCF depletion on phenotypes in living organisms compared to the minor immediate changes to gene expression observed in cultured cell lines will be essential in understanding the role of this proteins in transcription.
In addition to CTCF, other proteins present on chromatin may also affect progression of the cohesin ring and thereby influence chromatin interactions. Experiments measuring translocation of cohesin in vitro find that the process is hindered by DNA-bound proteins, including nucleosomes51,52. The degree of interference with translocation is directly related to the size of the protein or complex, likely due to the difficulty in passing through the cohesin ring51. This suggests that large complexes of transcription factors could present barriers to loop extrusion where the cohesin ring slows down, but not to the same extent as CTCF sites. This may partially explain enrichment of interactions within the interior of CTCF loop domains.Although the cohesin ring may form a relatively stable interaction with CTCF, the extrusion process is likely recurrent, with several rings constantly extruding along the same loop47. This concept has important mechanistic consequences for gene expression. Continuous extrusion results in frequent interactions between sequences within the loop, contributing to the intensity of Hi-C signal in the interior of loop domains. Although these interactions are in principle random, transient retention of cohesin at sites of large protein complexes may help bring these sequences together and increase their interaction frequency. For example, large protein complexes bound to enhancers or promoters, such as Mediator or protein complexes bound to histone modifications, may help enhancers and promoters located within the loop contact each other. The extrusion-dependency of this process may explain the enhancer dependent bursting of transcription activation as genes are activated each time cohesin-mediated extrusion brings promoters in contact with enhancers88 (Figure 4B). However, this model fails to explain why acute depletion of RAD21 only has a minor effect on gene expression. Recently, it was found that many enhancer-promoter interactions are dependent on Yin Yang 1 (YY1)89,90. These enhancers are enriched with both YY1 and cohesin, and it is thought that YY1 may partially block cohesin mediated loop extrusion to form enhancer-promoter interactions88–90. Interestingly, YY1 is able to dimerize and enhance DNA interactions in vitro without requiring cohesin89. Therefore, it is possible that cohesin-mediated extrusion may initiate enhancer-promoter interactions that are then partially stabilized by YY1 dimerization. This may explain the minor effect that acute cohesin degradation has on gene expression. A deeper exploration of this idea and the characterization of other proteins that are involved in blocking loop extrusion or in stabilizing enhancer-promoter interactions will be important in future work.
Interactions among A or B compartmental domains may also help stabilize active or silenced transcriptional states. Contacts among A compartments presumably take place through interactions among multivalent proteins present at enhancers and promoters, as well as RNAs and components of the splicing and termination machinery. These interactions may contribute to the co-regulation of genes bound by similar transcription factors and to an increase in the local concentration of the transcription machinery, resulting in the formation of structures similar to transcription factories. Interestingly, these structures are not stable, as they must be disrupted by the continuous extrusion via the cohesin complex, which increases interactions within loops while decreasing contacts between loops and compartmental domains.
Conclusion and Future Perspectives
Results discussed here suggest that the genomes of all organisms examined to date are organized into compartmental domains. These domains may represent the most basic form of 3D chromatin architecture and they are established as a consequence of interactions among protein complexes associated with DNA sequences based on their transcriptional or chromatin state. In vertebrates, an additional level of organization is established as a result of the extrusion process mediated by cohesin and perhaps also condensin. Stalling of extrusion by CTCF leads to the formation of stable loops that regulate enhancer-promoter interactions. It is possible that a subset of CTCF loops are common to all cells and are maintained in the germline and early embryogenesis, in which case the resulting 3D architecture imposed by these loops could be considered to regulate transcription, rather than being a consequence of this process. However, CTCF is present at many sites in the genome that lack the CTCF motif, where it may be recruited by transcription factors, in which case the organization imposed by these loops would be a consequence of transcription. In addition to further exploring the relationship between 3D genome organization and transcription, improvement in the following three technical areas may lead to important advances the field:
Shrinking Genome Organization.
Improvements to the Hi-C methodology and a lower cost of sequencing have enhanced the resolution at which chromatin organization can be visualized. Continued improvements in Hi-C resolution may allow the understanding of the contribution of single genes or regulatory sequences to 3D chromatin organization. Hi-C maps also suggest the existence of simultaneous interactions among multiple loci in the genome, but this may be a consequence of the cell populations used to obtain most Hi-C datasets. Technical innovations that allow the sequencing of long reads should afford the visualization of possible multi-loop structures contributing to 3D organization and their functional significance.
Visualizing Loop Extrusion.
The extrusion process has not been directly measured or visualized for cohesin in mammalian cells. Full acceptance of this model will depend on obtaining direct evidence for the extrusion process in the context of CTCF loops in vivo. Although work in vitro and in bacterial systems using similar SMC complexes is promising, further work with mammalian cohesin on actual chromatin will be an important step forward.
Population versus Single-Cell Chromatin Organization.
Information from thousands of single-cell Hi-C maps have been used to track the dynamics of CTCF loops and compartmental domains. The results suggest that features of chromatin organization may vary significantly between individual nuclei6. However, single-cell Hi-C studies have been limited by the coverage, and therefore resolution, achievable for these Hi-C maps2,6,92. Microscopy methods such as Oligopaint with STORM imaging are approaching the resolution at which domain structures can be visualized in single cells4. Thus, a combination of genomics and microscopy approaches may be useful in examining single-cell chromatin organization.
Key Points.
Results from high-resolution Hi-C in cells depleted of architectural proteins suggest two partially independent features of the 3D genome: compartmental domains and CTCF loops.
Compartmental domains are formed by interactions among multivalent proteins present at regulatory sequences of genes according to their transcriptional or chromatin state.
CTCF loops may be established as a consequence of continuous extrusion by multiple cohesin complexes along the 10 nm chromatin fiber. Loop extrusion is a pervasive, energy-driven process that may be also responsible for other aspects of transcription.
The functional output and 3D architecture of the genome are closely interrelated and reciprocally affect each other.
Acknowledgments
Work in the authors’ lab is supported by U.S. Public Health Service Award R01 GM035463 (V.G.C.) and Pathway to Independence Award K99/R00 GM127671 (M.J.R.) from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Glossary Terms
- ChIP-exo / ChIP-nexus
Chromatin Immunoprecipitation followed by exonuclease digestion. This technique is sed in place of standard ChIP-seq to identify protein binding sites at higher resolution. ChIP-nexus utilizes a different library preparation strategy to reportedly improve signal compared to ChIP- exo.
- CTCF loops
Point-to-point interactions between loci that coincide with CTCF and cohesin occupancy and often containing CTCF motifs in convergent orientation. These appear as bright punctae corresponding to high frequency interactions in Hi-C contact maps.
- GRO-seq
Global run-on sequencing is a method involving isolation of nascent transcripts and high- throughput sequencing to study active transcription genome-wide.
- Hi-C
A method using proximity ligation and high-throughput sequencing to identify all interactions taking place throughout the genome.
- Loop extrusion
A model in which chromatin is pulled through the cohesin or condensin ring to form loops.
- Oligopaint
A method of labeling DNA using short fluorescently labeled oligos for high resolution imaging of chromatin.
- STORM
Stochastic optical reconstruction microscopy. Super resolution imaging using individual photo- switchable fluorophores.
- Transcription Factory
Distinct nuclear location where RNAPII accumulates based on the observation that components of the transcription complex can be detected as discrete foci by microscopy. The transcription factory hypothesis suggests that genes are recruited to these nuclear locations in order to be transcribed.
- Transcriptional state
The state of a locus based on the presence chromatin-bound proteins or covalent histone modification that correlate with gene silencing or active transcription.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Fraser J, Williamson I, Bickmore WA & Dostie J An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol. Mol. Biol. Rev. MMBR 79, 347–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevens TJ et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagano T et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beliveau BJ et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun 6, 7147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni Y et al. Super-resolution imaging of a 2.5 kb non-repetitive DNA in situ in the nuclear genome using molecular beacon probes. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagano T et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pegoraro G & Misteli T High-Throughput Imaging for the Discovery of Cellular Mechanisms of Disease. Trends Genet. TIG 33, 604–615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman-Aiden E et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou C, Li L, Qin ZS & Corces VG Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol. Cell 48, 471–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sexton T et al. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 148, 458–472 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Nora EP et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonev B & Cavalli G Organization and function of the 3D genome. Nat. Rev. Genet 17, 661–678 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Schmitt AD, Hu M & Ren B Genome-wide mapping and analysis of chromosome architecture. Nat. Rev. Mol. Cell Biol 17, 743–755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hnisz D, Day DS & Young RA Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell 167, 1188–1200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merkenschlager M & Nora EP CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genomics Hum. Genet (2016). doi: 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- 17.Ong C-T & Corces VG CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet 15, 234–246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lajoie BR, Dekker J & Kaplan N The Hitchhiker’s guide to Hi-C analysis: Practical guidelines. Methods 72, 65–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao SSP et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowley MJ et al. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell (2017). doi: 10.1016/j.molcel.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao S et al. Cohesin Loss Eliminates All Loop Domains, Leading To Links Among Superenhancers And Downregulation Of Nearby Genes (2017). doi: 10.1101/139782 [DOI] [Google Scholar]
- 22.Dong P et al. 3D Chromatin Architecture of Large Plant Genomes Determined by Local A/B Compartments. Mol. Plant 10, 1497–1509 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Haddad N, Jost D & Vaillant C Perspectives: using polymer modeling to understand the formation and function of nuclear compartments. Chromosome Res 25, 35–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Marco E, Pinello L & Yuan G-C Predicting chromatin organization using histone marks. Genome Biol 16, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Pierro M, Cheng RR, Lieberman Aiden E, Wolynes PG & Onuchic JN De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc. Natl. Acad. Sci. U. S. A 114, 12126–12131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Brandão HB, Le TBK, Laub MT & Rudner DZ Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le TBK, Imakaev MV, Mirny LA & Laub MT High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L et al. Widespread rearrangement of 3D chromatin organization underlies polycomb- mediated stress-induced silencing. Mol. Cell 58, 216–231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hug CB, Grimaldi AG, Kruse K & Vaquerizas JM Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 169, 216–228.e19 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Bensaude O Inhibiting eukaryotic transcription. Which compound to choose? How to evaluate its activity?: Which compound to choose? How to evaluate its activity? Transcription 2, 103–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Z et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232–235 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Ke Y et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 170, 367–381.e20 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Naumova N et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung YH et al. Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Rep 18, 1366–1382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Battulin N et al. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol 16, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cubeñas-Potts C & Corces VG Architectural proteins, transcription, and the three- dimensional organization of the genome. FEBS Lett 589, 2923–2930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cubeñas-Potts C et al. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res 45, 1714–1730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harlen KM & Churchman LS The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol 18, 263–273 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larson AG et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y, Currie SL & Rosen MK Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem jbc.M117.800466 (2017). doi: 10.1074/jbc.M117.800466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Lee R et al. Classification of intrinsically disordered regions and proteins. Chem. Rev 114, 6589–6631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brackley CA, Johnson J, Kelly S, Cook PR & Marenduzzo D Simulated binding of transcription factors to active and inactive regions folds human chromosomes into loops, rosettes and topological domains. Nucleic Acids Res 44, 3503–3512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Z et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 163, 1611–1627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Y et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanborn AL et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U. S. A (2015). doi: 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fudenberg G et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nichols MH & Corces VG A CTCF Code for 3D Genome Architecture. Cell 162, 703–705 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nasmyth K Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet 35, 673–745 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Alipour E & Marko JF Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res 40, 11202–11212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stigler J, Çamdere GÖ, Koshland DE & Greene EC Single-Molecule Imaging Reveals a Collapsed Conformational State for DNA-Bound Cohesin. Cell Rep (2016). doi: 10.1016/j.celrep.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson IF et al. Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J (2016). doi: 10.15252/embj.201695402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanke M, Tahara E, Huis In’t Veld PJ & Nishiyama T Cohesin acetylation and Wapl- Pds5 oppositely regulate translocation of cohesin along DNA. EMBO J 35, 2686–2698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ganji M et al. Real-time imaging of DNA loop extrusion by condensin. Science eaar7831 (2018). doi: 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terakawa T et al. The condensin complex is a mechanochemical motor that translocates along DNA. Science 358, 672–676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murayama Y, Samora CP, Kurokawa Y, Iwasaki H & Uhlmann F Establishment of DNA-DNA Interactions by the Cohesin Ring. Cell 172, 465–477.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagy G et al. Motif oriented high-resolution analysis of ChIP-seq data reveals the topological order of CTCF and cohesin proteins on DNA. BMC Genomics 17, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarzer W et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature (2017). doi: 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wutz G et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haarhuis JHI et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693–707.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gassler J et al. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 36, 3600–3618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu Y, Sinha M, Peterson CL & Weng Z The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome. PLoS Genet 4, e1000138 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mawhinney MT et al. CTCF-Induced Circular DNA Complexes Observed by Atomic Force Microscopy. J. Mol. Biol 430, 759–776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nora EP et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kubo N et al. Preservation of Chromatin Organization after Acute Loss of CTCF in Mouse Embryonic Stem Cells (2017). doi: 10.1101/118737 [DOI] [Google Scholar]
- 66.Phanstiel DH et al. Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development. Mol. Cell 67, 1037–1048.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonev B et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171, 557–572.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibcus JH et al. A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen AS, Pustova I, Cattoglio C, Tjian R & Darzacq X CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tran NT, Laub MT & Le TBK SMC Progressively Aligns Chromosomal Arms in Caulobacter crescentus but Is Antagonized by Convergent Transcription. Cell Rep 20, 2057–2071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brackley CA et al. Nonequilibrium Chromosome Looping via Molecular Slip Links. Phys. Rev. Lett 119, (2017). [DOI] [PubMed] [Google Scholar]
- 72.Flamholz A, Phillips R & Milo R The quantified cell. Mol. Biol. Cell 25, 3497–3500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vian L et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell (2018). doi: 10.1016/j.cell.2018.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ocampo-Hafalla M, Muñoz S, Samora CP & Uhlmann F Evidence for cohesin sliding along budding yeast chromosomes. Open Biol. 6, 150178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bausch C et al. Transcription Alters Chromosomal Locations of Cohesin in Saccharomyces cerevisiae. Mol. Cell. Biol 27, 8522–8532 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Busslinger GA et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Racko D, Benedetti F, Dorier J & Stasiak A Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res (2017). doi: 10.1093/nar/gkx1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jonkers I & Lis JT Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol 16, 167–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moore JM et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PloS One 7, e34915 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dowen JM et al. Control of Cell Identity Genes Occurs in Insulated Neighborhoods in Mammalian Chromosomes. Cell 159, 374–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wan L-B et al. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Dev. Camb. Engl 135, 2729–2738 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gregor A et al. De novo mutations in the genome organizer CTCF cause intellectual disability. Am. J. Hum. Genet 93, 124–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kemp CJ et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 7, 1020–1029 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katainen R et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet 47, 818–821 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Narendra V et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Wit E et al. CTCF Binding Polarity Determines Chromatin Looping. Mol. Cell 60, 676–684 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Zuin J et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci 111, 996–1001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fukaya T, Lim B & Levine M Enhancer Control of Transcriptional Bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weintraub AS et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588.e28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beagan JA et al. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res 27, 1139–1152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan X et al. YY1 controls Igκ repertoire and B-cell development, and localizes with condensin on the Igκ locus. EMBO J 32, 1168–1182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Flyamer IM et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte- to-zygote transition. Nature 544, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]