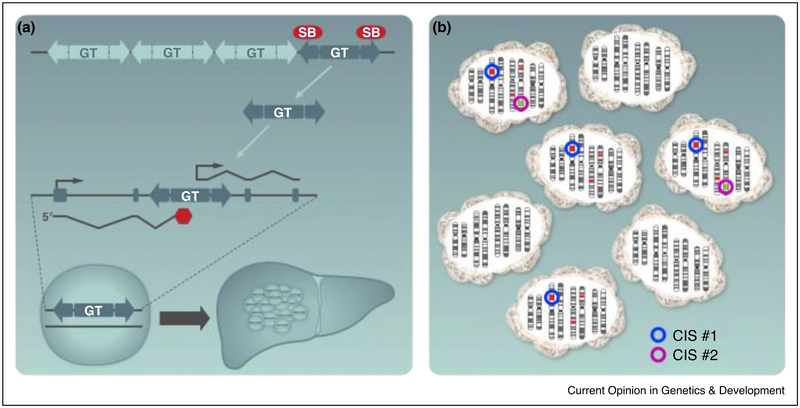
Figure 1. Transposon mobilization disrupts gene function by introducing gain-of-function or loss-of-function mutations.
(a) The SB and PB systems utilize two transgenic mouse lines, one harboring a concatemerized mutagenic gene trap (GT) that can disrupt gene function, and a second line carrying a ubiquitous or tissue-specific transposase that binds the transposon ends and catalyzes mobilization to new genomic sites. SB mutagenesis is depicted in liver cells, leading to the formation of liver tumors in vivo. The gene trap can alter gene function in two ways. In one or both orientations, a splice acceptor is followed by a polyadenylation (pA) signal. When the transposon inserts into a gene, the gene trap may splice to the transcript and the pA signal will prematurely truncate the mRNA, thereby disrupting expression of a candidate tumor suppressor. Additionally, a strong promoter followed by a splice donor (SD) is usually present in only one orientation. Transposon insertions that utilize this promoter/SD may drive overexpression of candidate oncogenes. (b) Bioinformatic analysis identifies common insertion sites (CISs) that represent genomic windows with more transposon insertions than predicted by chance. CISs 1 and 2 (represented as blue and purple circles) are found in independent tumors. Different methods of CIS identification are used to uncover genes that accelerate tumorigenesis, including Monte Carlo-based and Poisson distribution methods, Gaussian Kernel Convolution methods, and gene-centric common insertion site (gCIS) analysis.