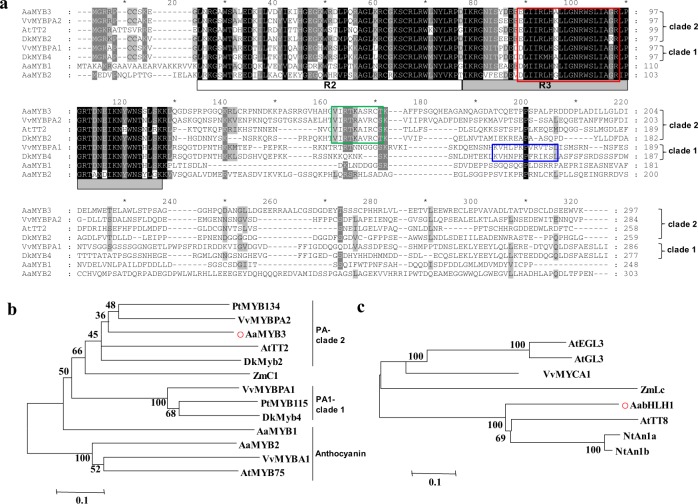
Fig. 1. Sequence alignments of AaMYB3, phylogenic tree of AaMYB3 and AabHLH1 with known R2R3-MYBs and bHLHs involved in proanthocyanidin (PA) biosynthesis.
a Alignment of the full-length deduced amino acid sequence of AaMYB3 with the other R2R3-MYBs. The R2 and R3 domains are shown; the bHLH interacting motif are indicated in the red box; the conserved motifs VI[R/P]TKAx1RC[S/T] for the PA-regulating MYBs PA-clade 2 are indicated in the green box, and the conserved motifs K[I/V]x2PKPx1Rx2S[I/L] for PA-clade 1 are indicated in the blue box. Identical nucleotides are shown on a black background, and gaps are indicated by dashes. b Phylogenetic relationship of AaMYB3 with selected known PA and anthocyanin MYB regulators from anthurium and other species. The scale bar represents 0.1 substitutions per site. c Phylogenetic relationship of AabHLH1 with selected known bHLH regulators involved in PA and anthocyanin biosynthesis and trichome development from other species. The scale bar represents 0.1 substitutions per site. The MYB names and GenBank accession numbers are as follows: AaMYB3, MH349476, AaMYB1, AAO92352.1, AaMYB2, AML84515 (Anthurium andraeanum); PtMYB134, ACR83705.1, PtMYB115, XM 002302608.2 (Populus spp.);VvMYBPA1, CAJ90831.1, VvMYBPA2, ACK56131.1, VvMYBA1, BAD18977.1 (Vitis vinifera): AtTT2, CAC40021.1, AtMYB75, NP 176057.1 (Arabidopsis thaliana); DkMyb2, AB503699, DkMyb4, AB503701 (Diospyros kaki); ZmC1, 1613412E (Zea mays); bHLH names and GenBank accession numbers are as follows: AabHLH1, MH349477; AtEGL3, AEE3412, AtGLABRA3, AED94664.1, AtTT8, AEE82802.1 (Arabidopsis thaliana); VvMYCA1, NP 001267954.1 (Vitis vinifera); ZmLc, NP 001105339.1 (Zea mays); NtAn1a, AEE99257.1, NtAn1b, and AEE99258.1 (Nicotiana tabacum)